How to Add New Line in Domain_9 Summary
| |
| What is a conserved domain? 3-D structures and Conserved features Domain family hierarchies |
| |
| CD-Search Results: Concise Display: top-scoring hits only  CD-Search Results: Standard Display: CD-Search Results: Full Display: all hits CD-Search Results: Small Triangles Specific Hits must meet or exceed |
| |
|
| ||||||||||||
![]()
![]()
![]()
![]()
Domains can be thought of as distinct functional and/or structural units of a protein. These two classifications coincide rather often, as a matter of fact, and what is found as an independently folding unit of a polypeptide chain also carries specific function. Domains are often identified as recurring (sequence or structure) units, which may exist in various contexts. The image below illustrates four "domains" identified as structural units in the MMDB-entry 1IGR, chain A, as segments colored in magenta, blue, brown, and green.In molecular evolution such domains may have been utilized as building blocks, and may have been recombined in different arrangements to modulate protein function. We define conserved domains as recurring units in molecular evolution, the extents of which can be determined by sequence and structure analysis.
Conserved domains contain conserved sequence patterns or motifs, which allow for their detection in polypeptide sequences. The distinction between domains and motifs is not sharp, however, especially in the case of short repetitive units. Functional motifs are also present outside the scope of structurally conserved domains. The CD database is not meant to systematically collect such motifs.
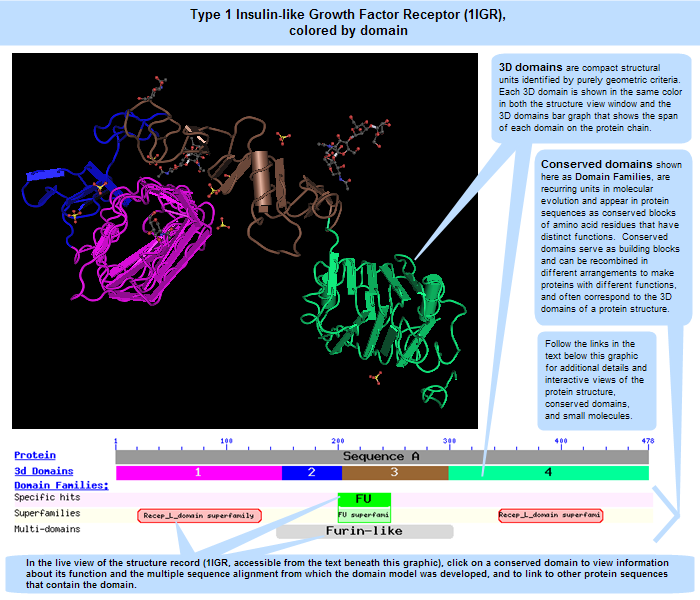 |
For this query sequence, a good correspondence exists between structural units (3D domains), identified by purely geometric criteria, and units asserted to be evolutionary conserved (domain families). The region annotated as "FU" (furin-repeat like) overlaps with a domain-split that was suggested by the MMDB domain parser. |
![]()
![]()
![]()
![]()
The two types of domains shown in the 1IGR illustration above -- 3D domains and conserved domains (or "domain families") -- often coincide with each other. However, because they represent two distinct types of data -- 3D structures and protein sequences, respectively -- they reside in two distinct databases: the Entrez Structure (Molecular Modeling Database, MMDB) and the Conserved Domain Database (CDD). The former includes the spatial (X,Y,Z) coordinates of each atom in a structure (where 3D domains are identified algorithmically), while the latter shows the span and composition of a conserved protein sequence region.Specifically, conserved domain models are based on multiple sequence alignments of related proteins spanning a variety of organisms to reveal sequence regions containing the same, or similar, patterns of amino acids. The illustration below provides an example, showing the multiple sequence alignment for the Furin-like domain, which is present in the Type 1 Insulin-like Growth Factor Receptor (1IGR) protein. Click anywhere on the image to open the complete, interactive CDD record for that domain model, cd00064. A separate section of this help document provides additional information about multiple sequence alignment display options.
In the CDD database, protein sequences from three-dimensional structures are included in domain models whenever possible, as one goal of the NCBI conserved domain curation effort is to make multiple sequence alignments agree with what we can infer from three-dimensional structure and three-dimensional structure superposition, in order to understand sequence/structure/function relationships. The sequence-based domain models and corresponding 3D structures are also cross-referenced to each other through Entrez "Links" between CDD and structure records.
 |
| This illustration shows the multiple sequence alignment for the Furin-like domain, which is present in the Type 1 Insulin-like Growth Factor Receptor (1IGR) protein. Click anywhere on the illustration above to open the live CDD domain summary record for CD00064: FU, Furin-Like Repeats and view the complete multiple sequence alignment for the Furin-like domain model. Separate sections of this CDD help document provide additional details about the source databases from which domain models are collected, the conserved domain assembly process, including the generation of a position-specific scoring matrix (PSSM) for each domain model, and multiple sequence alignment display options. |
![]()
![]()
![]()
![]()
Conserved Domains can be described by local multiple sequence alignments (illustration) spanning a variety of organisms to reveal sequence regions that contain the same, or similar, patterns of amino acids. Computational biologists from all over the world have compiled collections of such alignments representing conserved domains. CDD includes domains curated at NCBI as well as data imported from the external sources listed below, and data sources are indicated by their accession number prefixes.The source databases differ in their scope of coverage and the method by which they develop their models. Therefore, each source database may have its own model for a given conserved domain, in addition to some domain models found only in that database. To provide a non-redundant view of the data, CDD clusters similar domain models from various sources into superfamilies. The data sources include:
NCBI-Curated Domains



NCBI-curated domains use 3D-structure information to explicitly to define domain boundaries, aligned blocks, and amend alignment details. More details about the unique features of NCBI-curated domains are below.The goal of the curation project is to provide CDD users with insights into how patterns of residue conservation and divergence in a family relate to functional properties, and to provide useful links to more detailed information that may help to understand those sequence/structure/function relationships. The presence of conserved features help to affirm family membership in search results with borderline significance, for example. NCBI CDD Curators provide feature annotation and associated evidence in a computer friendly way, so that the scientific community can build software tools for the automation of tasks like annotation transfer, for example.
NCBIfams



NCBIfams is a collection of protein family hidden Markov models (HMMs) for improving bacterial genome annotation. A paper by Haft et al. (2018) provides additional information about NCBIfams, which is part of NCBI's Reference Sequence (RefSeq) project.
External Data Sources



In addition, CDD imports data from five other major sources, below. The version number (as available) of each source database that is imported into CDD is provided in the CDD News page.
Abbreviation Database Name Description SMART Simple Modular Architecture Research Tool SMART is a web tool for the identification and annotation of protein domains, and provides a platform for the comparative study of complex domain architectures in genes and proteins. SMART is maintained by Chris Ponting, Peer Bork and colleagues, mainly at the EMBL Heidelberg. CDD contains a large fraction of the SMART collection. Pfam Protein families Pfam is a large collection of multiple sequence alignments and hidden Markov models covering many common protein domains and families . Pfam is maintained by Alex Bateman and colleagues, mainly at the Wellcome Trust Sanger Institute. CDD contains a large fraction of the Pfam collection. COGs Clusters of Orthologous Groups of proteins COGs is an NCBI-curated protein classification resource. Sequence alignments corresponding to COGs are created automatically from constituent sequences and have not been validated manually when imported into CDD. TIGRFAMs The Institute for Genomic Research's database of protein families TIGRFAMs, a research project of the J. Craig Venter Institute, is a collection of manually curated protein families from The Institute for Genomic Research and consists of hidden Markov models (HMMs), multiple sequence alignments, Gene Ontology (GO) terminology, cross-references to related models in TIGRFAMs and other databases, and pointers to literature. PRK PRotein K(c)lusters Protein Clusters is an NCBI collection of related protein sequences (clusters) consisting of Reference Sequence proteins encoded by complete prokaryotic and chloroplast plasmids and genomes. It includes both curated and non-curated (automatically generated) clusters.
CDD also contains data from additional research projects, such as KOGs (a eukaryotic counterpart to COGs) and the Library of Ancient Domains ( LOAD ), contributed by I. Aravind, E. Koonin, and colleagues. The KOGs data set is accessible as a separate CD-Search database/Batch CD-Search database, and the LOAD data set is available on the FTP site, but neither of those data sets is directly searchable by text term in Entrez CDD.The content of imported domain models is determined by the providers of the source database, with slight modifications made at NCBI to link a domain model's member sequences to corresponding, complete protein sequence and 3D structure records in Entrez databases, when possible. The method by which imported domain models are integrated into the CDD database is described in the CD assembly process section of this help document.
Accession Prefixes indicate data sources:



Source databases are evident from CD accessions:
Accession starts with: Source Database cd Curated at NCBI sd
Domain models specifically built to annotate structural motifs;
this is a subset of the NCBI-curated domain models.
NF NCBIfams pfam Pfam smart SMART COG COGs KOG KOGs (available as a separate search set via CD-Search (RPS-BLAST); not searchable by text term in Entrez) PRK PRotein K(c)lusters (Entrez database) CHL
Chloroplast and organelle proteins; subset of the PRK database.
MTH
Mitochondrial proteins; subset of the PRK database.
PHA
Phage proteins; subset of the PRK database.
PLN
Plant-specific (non-chloroplast) proteins; subset of the PRK database.
PTZ
Protozoan proteins; subset of the PRK database.
TIGR TIGRFAMs LOAD_ Library of Ancient Domains (LOAD) data set. (available as a separate data set via FTP; not searchable by text term in Entrez)
Accessions that start with "cl" are for superfamily cluster records, which can contain domain models from one or more source databases.When searching CDD, it is possible to limit search results to domains from any given source database by using the Database Search Field.
![]()
![]()
![]()
![]()
NCBI-curated domain models are assembled using the methods briefly described in the source databases section of this document. More details about the NCBI curation process are provided by Marchler-Bauer, et al. (2007). An example of a multiple sequence alignment on which a model is based is shown in an illustration of the Furin-like domain.Domain models from external data sources are assembled by various methods, ranging from automated processing to manual curation, depending on the individual source database.
Upon import into CDD, protein sequence alignments (illustration) from each of the source databases are processed in an automated way to provide links from each aligned sequence to the corresponding, complete record in the Entrez Protein database. Occasionally, sequences that cannot be identified in Entrez's databases are omitted or substituted for closely related matches. Whenever possible, sequences in PFAM, SMART, and COGs alignments are substituted for closely related sequences (passing a stringent sequence similarity threshold) that have direct links to three-dimensional structures in the Moleclular Modeling Database (MMDB).A representative sequence is chosen for each domain model, preferably with a structure-link, for technical reasons. The representative sequence is generally shown as the first member of the multiple sequence alignment for a domain model. By default, this representative is the 3D structure shown when CD alignments are visualized with Cn3D.
A consensus sequence is computed from the imported alignments. Alignment columns have to be represented in at least 50% of all aligned sequences (weighted by diversity) to determine the extent of the consensus. The most frequently occurring residue in each column (after weighting to account for redundancy) is reported.
A position-specific scoring matrix (PSSM) is calculated for the extent of the consensus sequence. The PSSM profiles the various amino acids that were present in a given position of the multiple sequence alignment for a domain model and how frequently each one was observed. The consensus sequence does not contribute to the residue frequency statistics. Each PSSM receives a unique identifier (PSSM ID).
A PSSM ID is the unique identifier for a domain model's position-specific scoring matrix (PSSM). If a domain model's PSSM changes in any way as a result of updates to its multiple sequence alignment, it receives a new PSSM ID. This happens because a conserved domain model can evolve over time. For example, as new sequence data become available, the curators of a source database might add sequences to a multiple sequence alignment or update the sequences already present. As a result of such changes to the domain model, the PSSM and its ID can change. (Additional notes: Each superfamily record in the Conserved Domain Database also has a PSSM ID, which refers to the specific set of conserved domain PSSM IDs that comprise the superfamily, rather than to an actual position-specific scoring matrix for the overall superfamily. Obsolete PSSMs (e.g., 667) cannot be retrieved through the Entrez CDD search interface because they are no longer indexed. However, they can be retrieved from the archival copy of the database by using the "Direct Fetch via UID" option on the CDD Search Methods page.)
Search databases compiled of these PSSMs are available through the CD-Search service (see help document) and on the NCBI FTP site as collections of pre-computed RPS-BLAST databases that can be used for locally installed versions of that program.
![]()
![]()
![]()
![]()

As noted in the section on CDD data sources, NCBI-curated domains use 3D-structure information to explicitly to define domain boundaries, aligned blocks, and amend alignment details.
The goal of the NCBI conserved domain curation project is to provide database users with insights into how patterns of residue conservation and divergence in a family relate to functional properties, and to provide useful links to more detailed information that may help to understand those sequence/structure/function relationships. To do this, CDD Curators include the following types of information in order to supplement and enrich the traditional multiple sequence alignments that form the foundation of domain models:
3-dimensional structures and conserved core motifs: NCBI Conserved Domain Curators have re-evaluated and modified multiple sequence alignments imported from outside sources, and made them agree with what we can infer from three-dimensional structure and three-dimensional structure superposition. Curated alignments contain aligned blocks spanning all rows (with no gaps allowed inside blocks) and unaligned regions between blocks. The blocks are meant to represent conserved structural core motifs of the corresponding domain family. The 3D structures can be viewed interactively with the Cn3D structure viewing program. More information about viewing structures is provided in the section of this document on CD summary pages, and the illustration at the right provides an example of a protein structure that has been annotated by NCBI curators to highight the Cl- binding residues.
(Click on the illustration to open the current, interactive record for the Voltage-Gated Chloride Channel domain model, cd00400, in the Conserved Domain Database (CDD). From there, you can open an interactive version of the 3D structure, with conserved feature annotations, in the free Cn3D structure viewing program.)Conserved features/sites: In addition to working on the alignment model (illustration) , NCBI curators also record, when possible, the location and nature of features conserved in the domain family. Typically these would describe catalytic residues, binding sites, or motifs commonly referred to in the literature.

Features are added if they seem applicable to the family described in the CD's scope and if there is evidence linking the feature to a set of addresses on the alignment. Such evidence is recorded and available for inspection; it may be free-text comments, citations linked to PubMed, or "structure evidence" - exemplifying the existence of a site by highlighting an actual molecular complex, for example. Both features and evidence can be visualized on CD summary pages (in the conserved features/sites summary box, and as hash marks (#) in the multiple sequence alignment displays), and with the Cn3D structure viewing program. An example is shown in the illustration at the right. (Click on the illustration to open the current, interactive record for the Voltage-Gated Chloride Channel domain model, cd00400, in the Conserved Domain Database (CDD). Note that the live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data; however, the concepts shown in the illustration remain stable.)
In addition, the CD-Search tool can be used to identify conserved features in a query protein sequence, designated by small triangles (illustrated example) in the
search results graphical summary, when such features can be mapped from the conserved domain annotations to the query sequence.
Phylogenetic organization: Based on evidence from sequence comparison, NCBI Conserved Domain Curators attempt to organize related domain models into phylogenetic family hierarchies ( illustrated example ). The CDTree program used by NCBI curators can be downloaded in order to view NCBI-curated domains interactively and in greater detail.
Links to electronic literature resources: NCBI curated domains also provide links to citations in PubMed and NCBI Bookshelf that discuss the domain. These references are selected by curators and, whenever possible, include articles that provide evidence for the biological function of the domain and/or discuss the evolution and classification of a domain family.
NCBI-curated domains can be recognized in CDD search results by their "cd" accession number prefix. It is also possible to limit CDD search results to domain models from any given source database by using the Database Search Field.
![]()
![]()
![]()
![]()
A domain family hierarchy is a set of related domains that share a common ancestor, a common set of conserved residues, and a common general function, but differ from each other in their specific phylogeny, specific functions, and additional spans of conserved residues. Domain hierarchies are present in NCBI-curated domains in order to provide insights into how patterns of residue conservation and divergence in a family relate to functional properties.Some domain families have only a single node, while others have a hierarchy that is two or more levels deep, sometimes with numerous nodes ("subfamilies")at each level. Such hierarchies have generic "parent" models and more specific "children". The parent node contains a span of conserved residues that is also present in each of the children. Each of the child nodes can have additional conserved residues that extend beyond that span and help to further characterize the members of the child node.
NCBI CDD Curators attempt to split "children" nodes where they see evidence for ancient gene duplications resulting in orthologous groups, often occurring together with functional divergence. The CDTree program used by NCBI curators can be downloaded in order to view NCBI-curated domains interactively and in greater detail, with or without a query sequence embedded.
An illustrated example of a subfamily hierarchy is provided below.
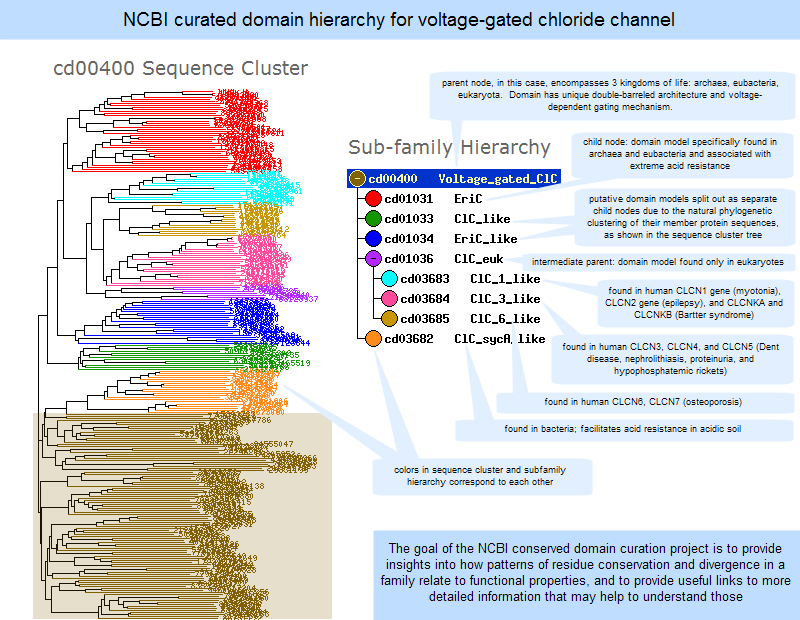
Click anywhere on the image to open the current, interactive record for the Voltage-Gated Chloride Channel domain model, cd00400, in the Conserved Domain Database (CDD). Note that the live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data; however, the concepts shown in the illustration remain stable.
![]()
![]()
![]()
![]()
A superfamily cluster is a set of conserved domain models that generate overlapping annotation on the same protein sequences. These models are assumed to represent evolutionarily related domains and may be redundant with each other. A superfamily accession number begins with the prefix "cl" for "cluster". (Some superfamilies contain only a single conserved domain model (singleton), and these are not indexed in Entrez. Only superfamilies that contain two or more conserved domain models are indexed in Entrez and will therefore appear in search results.)
Clustering methodology:Superfamily members are clustered through an automated process that involves the following steps:
- Identify domain models that have overlapping hits on sequences in the Entrez Protein database from at least five different identical protein groups (IPGs).
Technical note: In the data processing pipeline at NCBI, protein sequence records that contain an identical sequence, regardless of TaxID, are placed in an identical protein group (IPG), and each group is given a stable unique identification number (referred to as IPG ID or UID).- Store the overlapping domain models as pairwise associations, and use those pairwise associations to populate a similarity matrix.
- Refine the similarity matrix by comparing it against a "blacklist" (to remove unacceptable pairwise associations), and against a "whitelist" (to add pairwise associations that known to be valid but are not yet listed):
- The "blacklist" is used to separate domain models that should never be paired. The blacklist overrides all other aspects of the clustering algorithm.
For example:These pairings above are forbidden because the PA (protease-associated) domain is often inserted in a protease domain, yet the proteases that contain the insert are distinct from each other. For example, if a PA domain is in a serine protease and also in a metalloprotease, the two types of protease would be clustered together by the algorithm in the absence of a black list. However, the metalloprotease and serine protease are actually distinct, and simply represent convergent evolution to similar function.
- cd00538 (PA domain hierarchy) is blacklisted against pfam00082 (a serine protease model, subtilase family)
- cd00538 (PA domain hierarchy) is blacklisted against cd08022 (an M28-family metalloprotease)
- The "whitelist" is used to add pairwise associations of domain models that are known to be related, but that might not have been listed in the initial, unrefined similarity matrix.
For example:
- NCBI-curated domain models that are organized hierarchically are part of the white list.
- Conserved domain models from external databases can also be grouped together, if those domains are known to be related but were not grouped automatically by the clustering algorithm. An example of such a whitelisted pair is shown in cluster cl23875: MvaI_BcnI Superfamily, which includes pfam15515 (MvaI/BcnI restriction endonuclease family) and pfam09562 (LlaMI restriction endonuclease), as of 30 January 2018. Those two conserved domain models for restriction endonucleases (REs) have relatively few hits in common. RE superfamilies are very sequence-diverse, meaning that models for specific subfamilies can differ quite a bit in terms of overall length and conserved residue patterns/signatures. Nevertheless, the models are whitelisted together because they are known to be related.
- Take the refined similarity matrix and feed it to the Python "fastcluster" package (https://pypi.python.org/pypi/fastcluster), to create clusters using the "complete linkage" algorithm.
- Implement a post-processing step to compare the Python-generated clusters against the whitelist, in case Python did not put two domain models in the same cluster but should have.
NOTE: Multi-domain models that were computationally detected are not included in Superfamily clusters. These models are likely to contain multiple single domains and might falsely join superfamily clusters.
Rationale:
Superfamilies provide a method for organizing data within CDD in a non-redundant way. CDD contains conserved domains from a number of different source databases, each of which may have its own model for a given conserved domain. The models might share many similiarities in their reported residue conservation patterns, but differ in the specific protein sequences used in the multiple alignment, their footprint length [domain boundaries], and biological annotations. Because of the similarities, RPS-BLAST might find that multiple domain models align to the same general region of a query protein, but have different footprints and E-value scores relative to the query protein. If the footprints of two or more domain models overlap on the query, those models are clustered into the same superfamily, then the superfamily continues to be extended using the methodology described above.
Example:
One example of a superfamily is Cluster ID cl02915, which contains various domain models for the voltage-gated chloride channel. Superfamily members include the NCBI-curated domain cd00400 and all members of that family hierarchy plus domain models from external resources.
Selection of Superfamily Representative:
A superfamily can contain one to many domain models. As of spring 2008, approximately 70% of the ~9,000 superfamilies contain a single model and the rest contain multiple models. Single model superfamilies often represent proteins specific to certain organisms or taxonomic lineages (for example, viruses). The numbers of superfamilies containing single or multiple domain models will continue to evolve as new domains are imported and new NCBI-curated hierarchies are added.In superfamilies contatining multiple domain models, one of the models is selected as the source of the superfamily name and description. The representative is one of the following, listed in priority order:
- the parent node of an NCBI-curated domain family hierarchy, if one is present in the superfamily cluster. In the few cases where a superfamily contains more than one NCBI-curated domain, the parent of the hierarchy with the largest number of sequence hits is chosen as the superfamily representative.
- the Pfam domain model that hits the largest number of Entrez protein sequences in an RPS-BLAST search
- the SMART, COG, PRK, or CHL model that hits the largest number of Entrez protein sequences in an RPS-BLAST search
- the sole member of a superfamily
Superfamily can change over time:
The composition of a cluster can change over time due to a variety of factors, such as:A superfamily cluster accession number will remain the same if at least 50 percent of its member models (conserved domain accessions) have not changed relative to the previous version of the cluster.
- availability of new domain models as the Conserved Domain Database continues to grow
- changes to previously existing models
- new and/or updated sequence records in the Entrez Protein database
- refinements to the automated clustering procedures
If more than 50 percent of the conserved domain accessions from a previous version of a cluster are no longer present in the new build of that cluster, or if the cluster size more than doubles with a new build, then the superfamily cluster accession is retired and replaced by a new accession(s). If two previous clusters merge into a single new cluster, the superfamily cluster accession number of the larger component cluster is used for the new grouping.
A superfamily also has a PSSM ID, which refers to the specific set of PSSM IDs for the domain models that comprise the superfamily, rather than to an actual position-specific scoring matrix for the overall superfamily. The superfamily PSSM ID will change if there is any change to the set of member PSSM IDs relative to the previous version of the cluster (e.g., if a member conserved domain gets a new PSSM ID due to changes in its multiple sequence alignment, of if a new conserved domain model is added to the superfamily as the result of a CDD database update).
The CD summary page for a Superfamily record
does not include a multiple sequence alignment display; rather, it provides the name and description of the superfamily and lists the domain models that belong to it. The multiple sequence alignment for any member domain model can be viewed by clicking on it to open its CD summary page.Superfamilies that contain a single domain model ("singletons")
The concept of superfamilies was applied to CDD in order to cluster related conserved domain models together and provide a non-redundant view of the available domain models. After the superfamily clustering algorithm is applied to the domain models in CDD, all resulting clusters are referred to as superfamilies, regardless of how many domain models they contain. The non-redundant view of CDD therefore includes superfamilies with a single domain model ("singletons") as well as superfamilies containing two or more domain models.
In the user interface, however, superfamilies that contain only one model are not displayed in search results, or listed as links from the domain model, because they look very similar to the model itself.
In contrast, superfamilies that contain two or more models ("multi-model superfamilies") are displayed in search results, and are also accessible as links from their member domain models. The number of multi-model superfamilies is provided in the Database Statistics box on the "Conserved Domains and Protein Classification News" page, and they can be retrieved by clicking on that statistic.
![]()
![]()
![]()
![]()
| Protein query sequence | Text term search in Entrez CDD | Protein → Conserved Domains links | Domain architecture |
Protein Query Sequence (CD-Search) :



Most users will explore conserved domains starting from CD-Search results for a protein of interest.The query can be a protein sequence in FASTA format or the GI or Accession of a protein sequence that exists in the Entrez Protein database.
The search results will show the conserved domains found in the protein. The colored bars that depict the domain footprints (shown in both the concise display and full display of CD-Search results) are active hotlinks that open the corresponding CD summary pages with your
query sequence embedded in the multiple sequence alignment of proteins used to create the domain model.The second half of this help document provides details on how to use the CD-Search service, including input required and output shown.
Text Term Search in Entrez CDD :



| allowable search terms | search methods: basic and advanced | search fields | quotes | wild card * |
| search results | document summary page | "display", "show, "sort by", "send to" menus) |
Allowable search terms



Conserved domains can be searched by text term in the Entrez CDD database. The Entrez query interface allows searching for keywords, publication dates, and taxonomic span, accesssion numbers, and more. The search field summary table in this document shows the variety of terms that can be used to query the database and provides sample searches. It is also possible to use quotes to force multiple terms to be searched as a phrase, and to use an asterisk (*) as a wild card to search for a word stem.For example, search the Entrez CDD database for strings like "Kinase" or "pfam023*" or "Tetratrico*" to see how it works:
A number of techniques can be used to search the database, offering varying degrees of control over your query. The search methods summary table provides examples of basic and advanced searches. In basic searches, you can just enter one or more search terms without specifying search fields, Boolean operators, or other search criteria. These searches are quick and easy but can result in some extraneous hits. Advanced search methods, on the other hand, allow you to exercise greater control over your search, for example, by specifying which search field to use for each query term, limiting search results to a particular type of record or source database, or refining your search in other ways. A separate section of this help document describes the CDD search results.(The PubMed help document and Entrez help document provide additional, general information about using the Entrez search system.)
Search Methods



A variety of techniques can be used to search the Entrez CDD database, offering varying degrees of control over your query. In some cases, they offer alternative ways of executing the same search (as is true for sample searches #4, #5, and #6 below), with each method offering different benefits. The search methods include:
- Basic search (& search details)
- Limits
- Advanced search (Search builder, Show index list, History)
- Complex Boolean query
- Batch query
Additional details about search methods and options are provided in the: (1) PubMed help document (including information about temporarily saving records from your search results to the Clipboard); (2) My NCBI help document (including information about Saving search strategies and indefinitely saving records from your search results into your My NCBI Collections); and (3) general Entrez help document. | ||||||||||||||||||||||||||
Search Fields



By default, the Entrez system searches all fields of a record. If you want to narrow your query by searching for your term(s) in a specific search field, you can select the desired field by using the pull-down menus on either the Limits and Advanced search page, or you can type the search field directly in your query (surrounding field names with square brackets [], for example, [Organism] or [Orgn]).* The Show index link on the Advanced search page allows you to browse the index of each search field, where you can see the available terms, the number of records containing each term or phrase, as well as the syntax for entering values in search fields such as Modification Date or Publication Date.The currently available fields include:
* In a query, the field name may be typed as the full name or abbreviation, and may be in upper, lower, or mixed case. It must be surrounded by square brackets []. A space between the search term and the field specifier is optional. If desired, surround a phrase with quotes to force an adjacency search. For example, the sample queries below will work equally: "chloride channel" [WORD] "chloride channel"[WORD] "chloride channel" [word] "chloride channel"[Text Word] ** The quotes surrounding the search terms in the All Fields example ensure the terms are searched as a phrase. If quotes are not used and the terms are not automatically recognized as a phrase by the Entrez system, Entrez will insert a Boolean AND between the terms and they may or may not appear adjacent to each other in the retrieved records. More search tips are provided in the PubMed help document and Entrez help document. It is also possible to search for a word stem by using an asterisk (*) as a wild card; for example, chlori* will retrieve records with terms such as chloride, chlorin, chlorinate, chlorite. The Entrez Help document provides additional information about truncating search terms in this way. |
Search Results



| document summary page | display settings: format, items per page, sort by | send to | filter your results | find related data |
Document Summary (DocSum) Page



After querying Entrez CDD by text term, the initial search results page (also referred to as the document summary, or "DocSum") provides a list of the conserved domain records that contain your search terms. The terms can appear in any field of the record, unless a search field was specified in the query.(Note: A separate part of this document describes the results of a search by protein query sequence using CD-Search.)Click on the accession number or thumbnail image of any record on the DocSum page to view its conserved domain (CD) summary page.
If desired, you can narrow your search by restricting the query to a search field of interest or adding more terms with a Boolean AND.
Alternatively, you can broaden your search by adding more terms (e.g., synonyms) to your query with a Boolean OR, or by following links to Superfamily Members.
Click on the image above to open the live search results page in CDD. Note that the number of items retrieved may be different than shown here because the Conserved Domain Database continues to evolve with the addition of new data. |
Display Settings:



The "Display settings" menu on acts upon all of the conserved domain records (default) in your search results, or on the subset you have selected with checkboxes. You can select items from multiple pages of the search results, if desired.
Format Summary -- a summary of all of the structure records (default) retrieved by your search, or for those you have selected with checkboxes, in HTML format. The information shown for each record may include the following, as available:
- Short name, which concisely defines the conserved domain
- Thumbnail image indicating if the conserved domain includes a protein sequence from a 3D structure.
If a 3D structure is included, the thumbnail will be a still graphic of the actual domain structure.
If no 3D structure is available for the protein family from which the domain model was created, the thumbnail icon will show a schematic of a multiple sequence alignment.- First 100 characters of the text summary, which provides a synopsis of biological function and salient features of the domain
- Accession number
- PSSMid
- A subset of links to additional information about the domain, including a "View in Cn3D" link that opens an interactive view of the domain's 3D structure in NCBI's free Cn3D structure viewing program and links to related data in other Entrez databases. (Note: The "Find Related Data" menu in the right margin of the search results page provides a complete list of links. That menu retrieves related data for all records (default) retrieved by your search, or for the subset of records you have selected with checkboxes.)
Summary (text) -- a summary of the records retrieved by your search, in plain text format. By default, all records from your search result are listed. If you are interested only in specific records, select their checkboxes, select the desired display settings, and press "Apply" to view only those records. The information shown for each record is the same as in the "Summary" format described above, but does not include the subset of links to additional information. UI List -- a list of the unique identifiers (UI's) for all of the conserved domain records (default) retrieved by your search, or for those you have selected with checkboxes. Items per page By default, 20 documents are listed per page. If desired, decrease (to a minimum of 5) or increase (to a maximum of 200) the number of documents displayed per page then press the "Apply" button. Sort by Search results are displayed in order of decreasing relevance with respect to the query. Many search fields have a score or rank associated with them; for example, the Title and Organism fields have a high rank, while the Description of Sites field has a lower rank. The presence of a search term in any one or more of the fields is scored accordingly by the search system, and the total score given to a hit is used in determining its relevance to the query and therefore its placement on the search results page. Additional options are available to sort records by descending or ascending order of Accession, Database, Modification Date, Number of Sites, PSSM Length, Publication Date, and Structure Representatives. A few of these sort options will cause certain types of records to cluster at the top or bottom of the search results, depending on whether ascending (up) or descending (down) order is chosen. For example, if you sort by:Number of Sites - NCBI-curated domain models will appear at the top of search results (if "sort by number of sites (down)" order is selected) because conserved features, such as catalytic or binding sites, are annotated only on those domain models.
PSSM Length - the superfamily records will appear at the bottom (if "sort by PSSM length (down)" is selected) because they do not have an actual Position Specific Scoring Matrix (PSSM). Rather, each member of a superfamily has a PSSM and corresponding PSSM ID. A superfamily's PSSM ID refers to the specific set of conserved domain PSSM IDs that comprise the superfamily, rather than to an actual position-specific scoring matrix for the overall superfamily.
Structure Representatives - NCBI-curated domain models will tend to appear at the top of search results (if "sort by structure representatives (down)" order is selected) because the curation process includes incorporation of protein sequences from resolved structures. Domain models from external data sources may also contain structure representatives. As noted in the section on data processing, sequences in PFAM, SMART, and COGs alignments are substituted, whenever possible, for closely related sequences that have direct links to three-dimensional structures in the Moleclular Modeling Database (MMDB).
Technical note: If you retrieve all records in the database by searching the Filter field for All[Filt], the records are simply displayed in descending order of UID (i.e., PSSM ID).
"Send To" menu options



The "Send To" menu options act upon all the hits retrieved by your search (default), or those you have selected by using their checkboxes.
File Saves all the hits retrieved by your search into a plain text file, in either "Summary (text)" or "UI List" format. Clipboard Copies all the hits retrieved by your search (default), or those you have selected with check boxes, into a Clipboard, which temporarily stores up to 500 items (they will be lost after 8 hours of inactivity). Click on the "Clipboard: XX items" link in the upper right corner of the page to view the items in any
format for up to 8 hours after your last activity in the database.The Clipboard will not add an item that is currently in the Clipboard; it will not create duplicate entries. You can remove items from the Clipboard, if desired.
Entrez uses cookies to add your selections to the Clipboard. For you to use this feature, your Web browser must be set to accept cookies.
Items in the Clipboard are represented by the search number #0, which may be used in Boolean search statements. For example, to limit the items you have collected in the Clipboard to those from human, use the following search: #0 AND human[organism]. This does not affect or replace the Clipboard contents.
The
Clipboard's "Send to" menu offers you the same "File" and "Collections" options as offered on the original search results page. The latter option saves all items (default), or the subset of items selected with check boxes, indefinitely in the My NCBI Collections section of your My NCBI account.Collections Saves all the hits retrieved by your search (default), or those you have selected by using their checkboxes, into the My NCBI Collections section of your My NCBI account.
Filter your results



The "Filter your results" area in the upper right corner of a search results page allows you to see all the records (default) retrieved by your search, or subsets of your search results that reflect commonly requested categories of records, and shows the corresponding number of records in each case.The links for "NCBI-curated," "imported," "
families " (individual conserved domain models), and "superfamilies" (clusters of evolutionarily related conserved domain models into which the individual conserved domain models fall) show the number of retrieved records that fall into each of those categories, and allow you to view those subsets of your search results, if desired.
Find related data:



The "Related information" box that appears in the right margin of the display for an individual record allows you to retrieve related data for that particular domain model. (For example, the "Related CDs/Superfamily Members" link for accession cd00400 will retrieve the other domain models in the Conserved Domain Database that appear to be evolutionarily related to or redundant with cd00400.)
A "Find Related Data" box (instead of an "Related information" box) will appear in the right margin of a CDD search results page if you retrieved two or more records. The "Find Related Data" box allows you to retrieve related data for all the models retrieved by your search (default), or for the domain models you have selected with checkboxes. (For example, the "Find Related Data" option for Related CDs/Superfamily Member Links will retrieve the other domain models in CDD that appear to be evolutionarily related to or redundant with the domains retrieved by your search, or with the domains you have selected with checkboxes.)
The links in either display can include the following, depending on the related data that are available for the domains you have retrieved:
A "Links" box also appears in the displays of individual conserved domain records. All links are described in the help document section on " CDD Record (CD summary page): What information is displayed for each domain model on its CD Summary page?" : "Links to related data in Entrez". The number and type of links that exist vary among CDD records, depending on the related data that are available for any given record.
- Related CDs
- Literature
- Sequence
- Structure
- BioSystems
- Other Links
Most links are accessible on both the search results page and on a CD summary page, although a few of the links are available in only one of those places (*), such as Representatives and Books links, which are available only on the CD summary page.
Entrez Protein links to Conserved Domains :



Another (indirect) way to search the Conserved Domain Database is to start in a database such as Protein or Structure and use the "Find Related Data" ad in the right margin of the search results page, or use the "Related Information" ad in the right margin of the display for an individual record, to traverse to conserved domains.For example, all sequence records in the Entrez Protein database have been RPS-BLASTed against the Conserved Domain database. These pre-calculated search results are available as "Conserved Domains" links from protein sequence records, making protein functional information one click away from the sequence record.
Several different types of links are available, allowing you to choose: (a) the format in which you want to view the conserved domains (e.g., in graphical format as domain footprints aligned to the protein sequence, or as a list of records from the Conserved Domain Database, each of which includes a multiple sequence alignment of the proteins used to create the domain model), and (b) the level of redundancy in the list of conserved domain models (e.g., a
concise list of the top scoring models or a full list of all models that have a statistically significant RPS-BLAST hit to the protein).The number of conserved domain models retrieved, and the order in which they are sorted/presented, depends upon the view you select:

Click on the image to open the actual Entrez protein sequence
record and follow the live links to conserved domains.
You may need to scroll down the display to see the links.CDD Search Results -- opens a graphical display (illustrated example) of conserved domain model footprints, ranked by their RPS-BLAST score and hit type. A model may appear more than once if it aligns to multiple regions of the query sequence. A concise display showing only the top-scoring hits is presented by default, and it can be changed to a full display of all hits if desired. more... Conserved Domains (Concise) -- opens a concise list of the conserved domain models that are the top-scoring RPS-BLAST hits to the protein query sequence. Each domain model is listed only once, even if a model had a hit to more than one region on the query sequence. more...
Conserved Domains (Full) -- opens a full list of all the conserved domain models that have a statistically significant RPS-BLAST hit to the protein. Each domain model is listed only once, even if a model had a hit to more than one region on the query sequence. more...
Domain Relatives -- opens a graphical display of similar domain architectures, as determined by the CDART tool, with links to the proteins that have each similar architecture. more...
More details about each link are provided in the table below:
Link Name What you will get: CDD Search Results The protein sequence → "CDD Search Results" link will open a graphical display, in the CD-Search tool, that shows conserved domain footprints aligned to the protein sequence, based on the results of an RPS-BLAST search of the protein sequence against the PSSMs of all the domain models in the Conserved Domain Database (CDD).
A concise display (depicted below and in a more detailed illustrated example), is shown by default and features only the top scoring models from various hit types. A toggle switch near the upper right corner of the display allows you to see the full display (illustrated example), if desired, which shows all domain models that meet or exceed the RPS-BLAST threshold for statistical significance, i.e., the E-value cutoff. (Open the protein sequence record GI 157830769, featured in the illustration below, and follow the "CDD Search Results" link to try it yourself.)
In both the concise and full views, small triangles indicate the amino acids involved in conserved feaures/sites, such as catalytic and binding sites, when such annotations are available in a domain model.
In these graphical views, the domain hits are ordered not only by E-value, but also by classification (specific hits, non-specific hits, superfamilies, multidomains) and model origin (with priority given to NCBI-curated domains if they meet or exceed a domain-specific threshhold score).
If a domain model aligns to more than one region of the query sequence, it will be listed multiple times in the list of domain hits. This is true because the alignment coordinates and score of the domain model vary among different regions of the query sequence, and each hit is reported separately. (The list of domain hits is not shown in the illustration above but is visible beneath the graphical summary on an actual CD-Search results page.)
Conserved Domains (Concise) The protein sequence → "Conserved Domains (Concise)" link will open a concise list, in the Entrez Conserved Domain Database, of the conserved domain models that were top-scoring hits to the protein query sequence. Each domain model will be listed only once, even if a model aligned to more than one region on the query sequence.
This list of domains corresponds to those shown in the concise
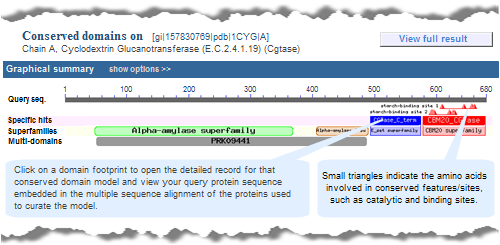
graphical display of CD-Search results. The same domains will appear in both views, but the order in which they are presented might vary between the displays because the graphical display sorts hits by RPS-BLAST score and hit type, and the list display does not. Also, if a superfamily on the "CDD Search Results" graphical display contains only one domain model , the "Conserved Domains (Concise)" link will retrieve the record for that sole domain model, rather than retrieving the superfamily record.For example, compare the concise set of top-scoring conserved domains on protein sequence GI 157830769, Cyclodextrin Glucanotransferase, in list vs. graphical format. Note that each link will open in a new window:
Conserved Domains (Concise) -- list format
CDD Search Results -- graphical display Conserved Domains (Full) The protein sequence → "Conserved Domains (Full)" link will open a full list, in the Entrez Conserved Domain Database, of the all the conserved domain models that have a statistically significant RPS-BLAST hit to the protein query sequence. Each domain model will be listed only once, even if a model aligned to more than one region on the query sequence.
This list of domains corresponds to those shown in the full
graphical display of CD-Search results. The same domains will appear in both views, but the order in which they are presented might vary between the displays because the graphical display sorts hits by RPS-BLAST score and hit type, and the list display does not. Also, if a superfamily on the "CDD Search Results" graphical display contains only one domain model , the "Conserved Domains (Full)" link will retrieve the record for that sole domain model, rather than retrieving the superfamily record.For example, compare the full set of top-scoring conserved domains on protein sequence GI 157830769, Cyclodextrin Glucanotransferase, in list vs. graphical format. Note that each link will open in a new window:
Conserved Domains (Full) -- list format
CDD Search Results -- graphical display, with "View Full Result" option activated Domain Relatives The protein sequence → "Domain Relatives" link opens a graphical display, in the Conserved Domain Architecture Retrieval (CDART) tool, of similar domain architectures, with links to the proteins that have each similar architecture . More information about CDART is provided below, in the "domain architecture" section of this document.
Domain architecture: CDART :



The Conserved Domain Architecture Retrieval Tool (CDART) program has been used to analyze the domain architecture of all sequence records in the Entrez Protein database, and to identify proteins with similar architecture. Those proteins are accessible by selecting "Domain Relatives" in the "Links" menu of a protein sequence record of interest ( illustrated example) .
Or, you can search CDART directly by entering a query protein sequence in FASTA format, or entering the GI or Accession number of a protein sequence that already exists in the Entrez Protein database. CDART will then retrieve proteins that contain one or more of the domains present in the query sequence.
More information about CDART is available in the overview, help document, and corresponding publication.
![]()
![]()
![]()
![]()
As you are viewing a search results page, click on the thumbnail image or title for any conserved domain model to see it's summary page. The thumbnail will show a snapshot of the 3D structure if a domain model includes a protein sequence(s) from a resolved 3D structure. The thumbnail will depict a sequence alignment if a domain model does not include protein(s) from a resolved 3D structure.A CD-summary page provides the following information for a domain model (example: cd00400: voltage-gated chloride channel):
- text summary (synopsis of function)
- links to the source database, literature references, and related data in Entrez, as available
- bioassay targets and results, as available
- statistics summarizing salient features of the domain model, such as number of protein sequence rows in the alignment, PSSM identifier, and more
- structure viewing options to display the 3D structure(s), if available, of protein sequences used to curate the domain model.
- conserved features (available for NCBI-Curated domains only)
- sequence cluster phylogenetic tree for protein sequences used to curate the domain (available for NCBI-Curated domains only)
- domain family hierarchy (available for NCBI-Curated domains only)
- multiple sequence alignments of the proteins used to develop the domain model.
More details about each section of the CD-summary page are provided below.
Text Summary (synopsis of function):



- The text summary shown at the top of a CD summary page was written by curators at the source database and provides a synopsis of the domain's biological function. In NCBI curated domains, it also describes the taxonomic extent of the domain, whether it is a monomer or dimer, and any salient features. The text summary in a superfamily record is derived from the representative domain.
Links to related data in Entrez:




- The "Links" box (illustrated at right) on an individual CD Summary page allows you to retrieve additional data associated with the domain model, including related models from the Conserved Domain Database, as well as literature, sequence, and other types of data from across the Entrez system.
- Most of the links are accessible from both the search results page (as "Related information" or "Find Related Data" ads in the right margin) and from the "Links" box on an individual CD Summary page. Links that are present on only one of the pages are noted with an asterisk (*), below. The number and type of links that exist vary among CDD records, depending on the related data that are available for any given record. The link types can include: Related CDs, Literature, Sequence, Structure, BioSystems, and Other Links.
- The "Source" link shown in the illustration opens either: (a) the root (parent) node of an NCBI-curated domain hierarchy, or (b) the corresponding domain record in the external source database, depending upon what type of domain model you are currently viewing. For example, if you are viewing a child or intermediate parent node of a hierarchy, the "Source" link will open the root node. If you are viewing the root node of the hierarchy, the "Source" link will open the corresponding domain model from an external database that was used as a seed for the curated model. If you are viewing the CDD display of a model that was imported from an external database, the "Source" link will open the corresponding domain record on the web site of the source database. In the latter case, note that CDD's display of the model might vary slightly from the source record due to the data processing procedures used by CDD.
Link Group Link Name * Description Related CDs Superfamily This links to the record for the CDD superfamily to which this domain belongs.
Superfamily Members This retrieves all the other domain models that belong to the superfamily.
Literature PubMed PubMed citations annotated on the domain. All references have been identified by curators, either by NCBI staff for the NCBI-curated domains, or by the staff of the external databases represented in CDD.
For NCBI-curated domains, the PubMed link leads to the citations that have been annotated on that particular node of a domain family hierarchy, not for all nodes in the tree. Whenever possible, the citations include articles that provide evidence for the biological function of a domain and/or discuss the evolution and classification of a domain family.
Free in PMC The subset of PubMed links that are available as free full text in PubMed Central.
Books * Full text information in the Entrez Books database that further clarifies or elucidates the domain's function, the protein's role in metabolic pathways, and other broad overview information, including diagrams and illustrations.
Sequence Representatives * The set of protein sequences that are present in the multiple sequence alignment for the domain model. Following the "Representatives" link will retrieve the complete sequence records from the Entrez Protein database. The number of records retrieved will be identical or similar to the number of Aligned Rows shown in the Statistics box of the CD Summary page.
Specific Protein The set of protein sequences found by RPS BLAST to contain the domain (with an E-value that is equal to or lower than a domain-specific Threshold E-value). These are called specific hits and represent a very high confidence that the query sequence belongs to the same protein family as the sequences use to create the domain model. The number of proteins you will retrieve by following this link is greater than retrieval from the "Representatives" link, but less than retrieval from "Related Protein" link.
Related Protein
(Protein †)Superset of all protein sequences found by RPS BLAST to contain the domain (with an E-value equal to or better than the default cutoff of 0.01). Therefore, this superset includes two CD-Search hit types: specific hits and non-specific hits.
Architectures
(Architecture †)Proteins found by CDART to contain one or more of the domains present in the proteins that are hit by domains found in the domain superfamily
Gene * Links from the RPS BLAST concise display hits to the protein sequences listed in Entrez Gene records.
Details: Each protein listed in an Entrez Gene record has been RPS BLASTed against the domain models in CDD. Links are then created between specific regions of those protein sequences and top-scoring domain models which align to them. Top-scoring domain models are shown either as specific-hits, or as the superfamily to which the highest-ranking non-specific hit belongs.
HomoloGene * Links from the RPS BLAST concise display hits to the protein sequences listed in HomoloGene records. (The details provided for Gene links, above, also apply to HomoloGene links.)
Structure Related Structure
(Structure †)Three-dimensional structures containing at least one protein molecule that has an RPS BLAST hit (specific hit or non-specific hit) to this domain model's PSSM (compare with "Family Structure").
BioSystems BioSystems * BioSystems containing proteins that have specific hits to the conserved domain. The proteins that have been associated with the BioSystem via the method descibed in data processing/create direct links/proteins.
Note that the BioSystems link can appear in two different places on a search results page:
- The "BioSystems" link in the Find Related Data ad in the right margin of the search results page will retrieve the biosystems associated with all of the conserved domains retrieved by your search, or with those you have selected with checkboxes
- The "BioSystems" link (when present) that is listed beneath an individual conserved domain record on a search results page (or in the "All links for this record" ad in the right margin of the display for an individual record) will open the subset of biosystems containing proteins annotated with that specific domain model. NOTE: If a conserved domain did not get any specific hits to proteins in any BioSystem, it will not have a BioSystem link.
Other Links Taxonomy The highest node in the NCBI Taxonomy database that encompasses all organisms whose protein sequences are in the multiple sequence alignment for a domain model. The taxonomy link for a superfamily retrieves the highest taxonomic node for all of its constituent domain models.
LinkOut * External data repositories can link to records in Entrez databases, including the Conserved Domain Database, by using the LinkOut tool. Selecting this option will display brief information about each conserved domain record you have retrieved (such as its accession and short name), followed by the names of (and links to) any external data providers that have chosen to link to the conserved domain (e.g., see cd05582, which has a LinkOut to "Domain Mapping of Disease Mutations").
Family Structure * Three-dimensional structures containing at least one protein molecule that has an RPS BLAST specific hit to this domain model's PSSM (compare with "Related Structure").
* Most of the links from conserved domain records to related data are accessible from both the search results page (as "Find Related Data" or "Related information" menus) and from the "Links" box on an individual CD Summary page. Links that are present on only one of the pages are noted with an asterisk (*) in the table above, specifically:The "representatives" and "books" links are found only on the CD summary page for a conserved domain; they are not present in the "Links" menus on a search results page.† Some links have a short name and a long name. The search results page often uses short names in its "Links" menus due to space limitations, such as "Protein" "Architecture," and "Structure." Those same links are called "Related Protein," Architectures," and "Related Structure," respectively, on an individual CD Summary page.Conversely, the "Gene," "HomoloGene," "LinkOut,"
"Family Structure," and "BioSystems" link currently appear only on the search results page.
BioAssay Targets and Results :



- A section entitled "BioAssay Targets and Results" appears on a conserved domain's summary page only if one or more members of the protein family have been used as targets in PubChem BioAssay records, and if at least one chemical was identified in the experiment(s) to be active against one of the targets defined by the domain family. As examples:
- view the conserved domain summary page for:
- cd09816: prostaglandin_endoperoxide_synthase (Animal prostaglandin endoperoxide synthase and related bacterial proteins)
- cd05061: PTKc_InsR (Catalytic domain of the Protein Tyrosine Kinase, Insulin Receptor)
- The "BioAssay Targets and Results" section lists bioassays that have tested the activities of small molecules (e.g., chemicals) against protein sequences that have a specific hit to this domain model, and are therefore considered to be members of this protein family. Some of the information from the bioassays may be generalizable to other members of the protein family, depending on how narrowly a family is defined.
- Up to three representative bioassays are listed as examples in the "BioAssay Targets and Results" box. Click on "Explore more" at the bottom of the box to see a complete list of experiments in the PubChem BioAssay database that have tested small molecules against protein targets belonging to this protein family. From there, you can open the "BioAssays" or "Compounds" folder tabs and click on the counts in the columns such as "active," "inactive," "tested" to see the chemicals that have been screened and their activity potency in the respective bioassays.
Statistics:



Item Description PSSM-ID the unique identifier for the position-specific scoring matrix (PSSM) generated by RPS-BLAST for a given multiple sequence alignment. If the sequence alignment changes in any way, for example, if new sequences are added to the alignment, a new PSSM will be generated and will receive a new PSSM-ID. (Note: Each superfamily record in the Conserved Domain Database also has a PSSM ID, which refers to the specific set of conserved domain PSSM IDs that comprise the superfamily, rather than to an actual position-specific scoring matrix for the overall superfamily.)
Aligned lists the number of rows in the sequence alignment . In general, each row comes from a different sequence record. However, sometimes two or more rows can be from the same GI number (i.e., same sequence record), if the sequence contains multiple instances of the domain. Threshold Bit Score the domain specific threshold score (shown as a bit score) that an RPS-BLAST hit must meet or exceed in order to be considered a specific hit, which represents a high confidence association between a protein query sequence and a conserved domain and therefore a high confidence level for the inferred function of the protein query sequence. The threshold is equal to the weakest E-value (and highest bit score) among self-hits of a domain�s member protein sequences to the resulting domain model (illustrated example). Domain-specific threshold scores are calculated only for NCBI-curated domains. Threshold Setting GI the GI number of the member protein sequence (i.e., the protein sequence from the domain model's multiple sequence alignment) that set the threshold bit score. A threshold setting GI number is displayed only for NCBI-curated domains, as thresholds are calculated as part of the curation process. Status information about the CD's curation status. Curated models have been realigned by NCBI with consideration of 3D structure. Alignments imported from outside sources have not been changed (except for the import process detailed above) Author name of the author who contributed the conserved domain model to the NCBI-curated data set. This line currently appears in records that were contributed by external collaborators (for example, cd08773). Mouse over the name to see a popup with additional contact information, if/as available, for the author. Created date at which the seed (or de-novo) alignment was imported into CDD Updated date of the most recent changes to the alignment model and/or descriptive information
Structure :



Item Description "Structure View" Button The "Structure View" button in a conserved domain record opens the 3D structure(s), if available, of protein sequences used to curate the domain model. In order for the button to work, the Cn3D program must be installed on your computer. It is a a free helper application available for Windows, Macintosh, and Unix platforms. Installation takes only a couple of minutes and a tutorial describes the program's features and functions. In addition to displaying an interactive view of the 3D structure(s), Cn3D will also display the multiple sequence alignment of those and other proteins used in the curation of the domain model. The Cn3D structure view and sequence view windows communicate with each other, so highlighting residues in one window will also highlight those residues in the other window.
As noted in the sections on the CD assembly process and unique features of curated domains, NCBI staff include protein sequences from resolved 3D structures
(illustration) whenever possible in the multiple sequence alignment of a domain model.In a multi-level domain hierarchy, the 3D structures might be present in the parent node (e.g., cd00400) if they are not present in an intermediate or terminal node (e.g., cd03683). In that case, click on the parent node to view structures that have been specially annotated to highlight the conserved feature.
You can click on any of the thumbnail structure images on a CD summary page to launch Cn3D. The thumbnail images in the conserved features summary box will launch a specially annotated view of the structure that highlights the particular feature of interest.
However, 3D structures are not always available. If a domain model does not include any structure-based protein sequences, the "Structure View" button will still open Cn3D, but only the sequence viewer window will be populated with data.
Controls in Cn3D will then allow you to manipulate the sequence alignment in various ways, if desired. For example, Cn3D offers column-specific coloring by sequence conservation when invoked with multiple alignment views. This is a convenient feature to study sequence conservation within a CD-alignment and to find out how well the aligned query fits the existing patterns of conservation and variability. The Cn3D tutorial provides more information on the controls available.
Program Although the Structure View button provides the option of using an older version of Cn3D (3.0), the default choice is recommended because it uses the most recent public version of the program (currently Cn3D 4.1). Drawing Structures, when available, can be displayed in varying levels of detail. All Atoms will load a detailed model. This option transmits a large amount of structure data and loading the structures may therefore take some time. The Virtual Bonds setting displays C-alpha atoms only, with virtual bonds connecting them, and therefore transmits and loads more quickly. Aligned Rows By default, Cn3D will display a multiple sequence alignment of up to 10 proteins, starting with sequences whose 3D structures are shown, and then also including sequences from proteins that do not yet have a resolved structure. Use the "aligned rows" menu to increase that number up to 100 rows.
Conserved Features/Sites summary box (available for NCBI-Curated domains only):



- If conserved features/sites have been annotated on an NCBI-curated domain, they are noted in a summary box near the top of the page, with one folder tab for each feature (illustrated example) .
- Click on the folder tab for a feature of interest to view its details, such as:
- feature number - The ordinal number of the feature within the conserved domain model (e.g., feature 1, feature 2, etc.)
- feature name - Generally reflects the function of the conserved feature/site (e.g., Cl- selectivity filter, Cl- binding residues [ion binding site], dimer interface [polypeptide binding site])
- conserved feature residue pattern - The set of amino acids that characterizes the conserved feature/site.
- The amino acids are not necessarily adjacent to each other in the domain model, but instead appear at the positions designated by hash marks (# symbols) in the multiple sequence alignment of the domain model, as described below.
- The pattern may include ambiguity codes (for example, [ST], which indicates that a position can be occupied by either Serine or Threonine).
- The conserved feature residue pattern is specified by curators, and displayed on a conserved domain summary page, only if it is clear that specific residue types are necessary for the particular molecular function (such as metal coordination, glycosylation attachment, or enzyme catalysis). Therefore, not all conserved features/sites will include a "conserved feature residue pattern" line.
- Note: If a sequence in the Entrez Protein database gets a significant hit to the conserved domain model AND contains the conserved feature residue pattern, the site annotation will be transferred to that protein sequence record.
- evidence - may be free-text comments, literature citations, or "structure evidence" that exemplifies the existence of a site by highlighting an actual molecular complex in an experimentally resolved 3D structure.
- 3D structure thumbnail image, if available. The conserved amino acids that characterize the feature are highlighted in pink, in both the thumbnail image, and in the larger, interactive view of the structure that appears when you click on the thumbnail to launch the structure in the free Cn3D viewing program. (Cn3D must first be loaded on your machine in order for that to work.)
- Clicking the folder tab for a feature of interest will refresh the mutliple sequence alignment display with an extra alignment row that shows the feature number (feature 1, feature 2, etc.) and uses hash-marks (#) to indicate the specific residues involved (also shown in the illustrated example ). Only one feature at a time is shown in the multiple sequence alignment display.
Sequence Cluster Phylogenetic Tree (available for NCBI-Curated domains only):



- Based on evidence from sequence comparison, NCBI Conserved Domain Curators attempt to organize related domain models into phylogenetic family hierarchies ( details and illustration ). Colors used in the sequence cluster phylogenetic tree correspond to colors used in the domain family hierarchy display. To examine the hierarchical classification more closely, you can download the CDTree program, which is used by the NCBI curators. CDTree enables you to interactively view the complete domain hierarchy, including a detailed display of the sequence cluster tree.
- To view a query protein embedded into the sequence tree of a domain model, first use the CD Search tool to identify the conserved domains in the query sequence. Then click on the cartoon (colored bar) representing a domain of interest in either the Concise Display or Full Display of the CD-Search results page. That will open a CD Summary Page, which shows detailed information about the domain and provides an Interactive Display option for viewing the Hierarchy (an illustrated example is provided in the "How To" pages).
To embed your query in the hierarchy, simply check the box for Add Query Sequence before pressing the "Interactive Display" button. (The free CDTree program must be loaded onto your computer in order for that button to work.) When the CDTree program opens, your query sequence will be highlighted in red. If the sequence tree is large, you might need to de-select the View/Fit to Screen option in CDTree's Sequence Tree window in order see the tree, and the placement of your query sequence, in detail. The CDTree help document is packaged with the software and provides details on how to use the program.
- Algorithms used to generate the cluster diagram in CDTree: The sequence tree viewer in CDTree calculates and displays sequence trees for a set of selected alignment models, which may or may not be linked in a hierarchical fashion. Sequence trees are the graphical depiction of results from simple phylogenetic analysis of the alignment data. Methods available for distance calculation are percent identity, Kimura-corrected percent identity, score of aligned residues, score of optimally extended blocks, blast score for the aligned footprints and blast scores for full-length sequences; a variety of commonly used scoring matrices can be selected. For the sequence trees displayed on CDD web pages, we commonly use "score of aligned residues", where pair-wise alignment scores derived from our multiple sequence alignments, and scored via BLOSUM62, are converted into distances. Trees can be constructed via single-linkage clustering, neighbor joining, or the Fast ME method. We use neighbor-joining for all of the sequence trees displayed on web-pages.
Domain Family Hierarchy (available for NCBI-Curated domains only):



- As noted in the description of NCBI curated domains, the goal of the curation project is to to provide CDD users with insights into how patterns of residue conservation and divergence in a family relate to functional properties. The CD summary page for an NCBI-curated domain shows the hierarchy ( details and illustration ) to which the currently viewed domain belongs.
Some hierarchies have only one node, while others have many nodes organized into two or more levels. If a hierarchy has multiple nodes, you can click on another node of interest to view the CD summary page for that domain.
Alternatively, you can download the CDTree program used by the NCBI curators in order to view the complete domain hierarchy interactively and in greater detail, with or without a query sequence embedded.
Multiple Sequence Alignment Displays :



- Member proteins used to create domain model:
By default, the sequence alignment display at the bottom of a CD summary page shows 10 of the most diverse members from the cluster of sequences used to create a domain model. (A sample multiple sequence alignment is shown in the illustration of cd00064: Furin-like domain in this help document, or you can open a domain model directly in CDD, such as cd00400: voltage-gated chloride channel.) The multiple sequence alignment display options (below) can be used to change the quantity and appearance of data displayed, and the CD-Search tool can be used if you'd like to embed a query sequence within the alignment.- Protein query sequence embedded in alignment:
To view a query protein embedded into the multiple sequence alignment of a domain model, first use the CD Search tool to identify the conserved domains in the query sequence. Then click on the cartoon (colored bar) representing a domain of interest in either the Concise Display or Full Display of the CD-Search results page.- Display Options:
By default, the multiple sequence alignment on a CD summary page is shown in hypertext format and displays up to 10 sequences that were used to curate the domain. The display format, number and type of sequence rows, and color scheme can be changed in the following ways:
Display Option Description Format
Hypertext Interactive view in which each accession or GI number links to the corresponding complete sequence record in the Entrez Protein database. Displays all residues in each sequence row, with aligned residues shown in upper case, unaligned residues in lower case, and variation in sequence length shown as dashes. A horizontal scale indicates the number of residues in the overall alignment. The numbers at the beginning and end of each sequence row indicate the span of sequence data that was imported from the complete protein sequence record.
Plain Text This view contains the same content as "Hypertext" but is rendered in ASCII format. Compact Hypertext Interactive view in which each accession or GI number links to the corresponding complete sequence record in the Entrez Protein database. Shows only aligned residues (as upper case letters), plus the number of intervening unaligned residues in each sequence row (shown in square brackets []). Does not show the unaligned residues themselves; those are shown only in the "Hypertext" and "Plain Text" format. Compact Text This view contains the same content as "Compact Hypertext" but is rendered in ASCII format. mFASTA Multiple FASTA (mFASTA) format is useful for importing the data into sequence analysis programs. For each sequence row in the alignment, it provides a FASTA-formatted definition line ("FASTA defline") followed by up to 80 characters of sequence data on each subsequent line. mFASTA format displays all residues in each sequence row, with aligned residues shown in upper case, unaligned residues in lower case, and variations in length filled in with dashes.
Row Display
Number of rows
in a domain modelThe total number of sequence data rows aligned in a domain model are shown in the statistics portion of that model's CD summary page.
Default number shown By default, 10 rows of sequence data are shown, including the representative sequence plus nine others. Maximum number shown You can change the number of sequence rows displayed using the Row Display pop-up menu. If the Type Selection is set to Most Diverse Members, a maximum of 100 rows can be displayed. If a domain model contains more than 100 rows, the Type Selection Top Listed Sequences allows the display of more than 100 rows. If a model is NCBI-curated, you can also use the CDTree program to view the complete set of rows. Simply install the program, which is free, then press the Interactive Display button in the hierarchy section of the domain model's CD summary page to view all the sequence rows. Note: In general, each row comes from a different sequence record. However, sometimes two or more rows can be from the same GI number (i.e., same sequence record), if the sequence contains multiple instances of the domain.
Type Selection
Most Diverse Members Lists the representative sequence followed by the most dissimilar protein sequences, as determined from the domain model multiple sequence alignment. They are listed from most to least dissimilar with respect to the representative sequence.
Top Listed Sequences Merely refers to the order in which the sequences are listed in the multiple alignment; this may or may not be meaningful, depending on the approach used by the source database in curating a particular domain model. In NCBI-curated domain models, protein sequences from resolved 3-D structures are generally listed first, so the "Top Listed Sequences" display option is useful for bringing these structure-based protein sequences to the top when viewing NCBI-curated domains. The remaining sequences in NCBI-curated domain models are listed in order of increasing GI number or some other non-biological criterion. (This is because the composition of the member sequences, not their order, is important in determining a domain model's position-specific scoring matrix, or PSSM. The other important factor is the degree of residue conservation in any given column of the alignment, which can be visualized with the Color Bits setting, described below.)
The biological relationships among the member sequences of an NCBI-curated domain model are displayed in the sequence cluster phylogenetic tree and the domain family hierarchy on the domain model's CD summary page. Both of these displays can also be viewed interactively using the CDTree program.
Color Bits
General Color Bits allow you to adjust the red <-> blue balance of color used to depict the degree of conservation among aligned (upper case) residues. In general, red indicates highly conserved and blue indicates less conserved. (In other words, the two extremes on the color scale correspond to columns that are completely conserved (e.g., same residue in all alignment rows), and columns with residue types distributed in a way that is no different from the background distribution -- what would seem like a random pick of residue positions from arbitrary protein sequences.) Unaligned (lower case) residues are shown in grey.
The color bit settings can be used to select a threshold
for determining which columns are colored in red .Numerical settings Higher numbers require higher degrees of conservation within an alignment column (i.e., less residue variation) in order to display that column in red font. The score threshold that must be met in order for an alignment column to be displayed in red can be adjusted from a low of 0.5 to a high of 4.0. As the threshold increases, the number of columns shown in red will decrease.
Background: Each column in the multiple sequence alignment display receives a score that indicates that column's "information content" -- its contribution to the overall alignment score -- indicating how important the column is as an "anchor" for the alignment. The higher the score, the more important that column is in the alignment.
We use a fairly standard definition of "information content" for an aligned column:
SUM (f(i) * log (f(i)/q(i)) over all base 2 residue types iwhere f(i) is the observed relative residue frequency, and q(i) is the background/reference relative frequency for that residue type (based on the table that accompanies the BLOSUM62 matrix). This is also called "relative entropy", which is a popular way to measure the distributions of nucleotide bases or amino acids.A column's score is calculated on the fly, based on the sequence rows currently shown in the display. As the number and type of sequence rows in the display change, the column's score, and therefore its color, can change.
Identity setting The Identity setting uses red font only in columns that contain the same residue in all of the sequence rows displayed. All other aligned columns are colored in blue and unaligned columns are shown in grey. "Feature" hash marks (#) Hash-marks (#) in the top row of a multiple sequence alignment display indicate the specific residues involved in a conserved feature, such as a binding or catalytic site, that has been annotated on an NCBI-curated domain.
Although multiple features may have been annotated, only one feature at a time is shown in the multiple sequence alignment display.
A conserved features/sites summary box
( illustration ) lists the features that have been annotated. Clicking on the tab for a feature of interest will show its details. It will also refresh the mutliple sequence alignment display to mark the residues involved in the currently viewed feature (as depicted in the bottom of the illustration ).
![]()
![]()
![]()
![]()
CDD is updated several times a year. We no longer try to follow updates of the source databases on a regular basis, but will re-import source database content occasionally. CDD continues to grow, however, through NCBI's curation effort. At the moment, CDD curators focus on capturing and describing hierarchies of related domain families, which are, for the most part, covered by the imported un-curated models as well. The current curation effort is restricted to ancient domain families with wide phylogenetic distribution, and focuses on families with at least one 3D structure representative.
![]()
![]()
![]()
![]()
The scientific community's understanding of molecular data continues to evolve as research progresses. Some domain models in CDD are generated through automated processes and others are curated. All are fluid and revised as new data become available and as new protein family clustering methods are developed. Because of this, we welcome your feedback on the data at info@ncbi.nlm.nih.gov, including information/annotations you find particularly helpful as well as any discrepancies you may notice.
![]()
![]()
![]()
![]()
The CD-Search service is a web-based tool for the detection of conserved domains in protein sequences. It can therefore help to elucidate the protein's function.The CD-Search service uses RPS-BLAST to compare a query protein sequence against conserved domain models that have been collected from a number of source databases, and presents results as a concise display (default), standard display, or full display.
If CD-Search finds a specific hit, there is a high confidence in the association between the protein query sequence and a conserved domain, resulting in a high confidence level for the inferred function of the protein query sequence. The other types of hits that can be found also shed light on the putative function of the query protein.
The CD-Search tool can also identify putative conserved features in a query protein sequence, when such features can be mapped from the conserved domain annotations to the query sequence. If conserved features are found, they designated by small triangles in the
search results graphical summary, indicating the specific amino acids likely involved in functions such as catalysis or binding.
![]()
![]()
![]()
![]()
The CD-Search service uses RPS-BLAST, which stands for "Reverse Position-Specific BLAST". This is a variant of the popular PSI-BLAST program ("Position-Specific Iterated BLAST"). PSI-BLAST finds sequences significantly similar to the query in a database search and uses the resulting alignments to build a Position-Specific Score Matrix (PSSM) for the query. With this PSSM the database is scanned again to eventually pull in more significant hits, and further refine the scoring model.RPS-BLAST uses the query sequence to search a database of pre-calculated PSSMs, and report significant hits in a single pass. The role of the PSSM has changed from "query" to "subject", hence the term "reverse" in RPS-BLAST.
RPS-BLAST is the search tool used in the CD-Search service. The CD-Search service provides a web-interface to the RPS-BLAST program, the CD search databases, and interactive alignment visualization including 3D structures. The search results can include several types of RPS-BLAST hits that represent various confidence levels (specific hits, non-specific hits) and domain model scope (superfamilies, multi-domains).
A standalone version of the RPS-BLAST program is available as part of the NCBI toolkit distribution. A separate section of this document describes the differences between the CD-Search web tool and the standalone RPS BLAST program.
![]()
![]()
![]()
![]()
| query sequence | options: database selection, expect value, Composition-corrected scoring, low complexity filter, force live search, rescue borderline hits, suppress weak overlapping hits, maximum number of hits, result mode | retrieve previous CD-Search result by RID |
Query Sequence :



You can submit a protein or nucleotide query sequence to CD-Search, either as a sequence identifier (i.e., as an accession or GI number that is valid in the NCBI Entrez system), or as FASTA-formatted or bare sequence data. Hitting the submit button will start CD-search with default settings for search sensitivity and display options.
Protein sequence queries: There is no length limit on a protein query (CD-Search does not check the length of a protein query sequence). Searches with multiple protein query sequences can be submitted to the Batch CD-Search tool, which also accepts input as either protein sequence identifiers or raw sequence data. The Batch CD-Search Help document provides details about formatting the input, including inserting line breaks between the query proteins. If multiple protein query sequences are entered in this way on the regular CD-Search page, your query will be automatically redirected to the Batch CD-Search tool. If there are no line breaks between the query proteins, however, an error message will be displayed and no redirect will occur. (Note that Batch CD-search only works for protein sequences; it does not accept nucleotide sequences.)Nucleotide sequence queries: The maximum length of a nucleotide query is 200,000 base pairs. The search system translates all 6 reading frames and scans the RPS-BLAST databases with the protein products. If a nucleotide query is input as a sequence identifier (accession or GI number), it will be translated using the genetic code that corresponds to the source organism of the sequence. If a nucleotide query is input as a FASTA-formatted or bare sequence data, it will be translated using the standard genetic code. Each translation is essentially processed like a separate protein query. In other words, 1 nucleotide sequence query = 6 protein queries. CD-Search will combine the results into a single page, but will only display the translated reading frames that picked up a match in CDD.
Options:



The options below are only available when using the actual CD-Search form. Searches launched from the CDD home page or together with protein BLAST requests use default search parameters. (The CDD home page does allow you to select the database, however.)
- Database Selection: currently, CD-Search is offered with the following search databases. Note that if you use the default "CDD" database, CD-Search automatically returns precalculated search results, unless you select the option to "force live search." If you select a database other than the default "CDD," the CD-Search program automatically uses the live search mode.
- CDD - this is a superset including NCBI-curated domains and data imported from Pfam, SMART, COG, PRK, and TIGRFAMs. It is the default database for searches.
- NCBI_Curated - NCBI-curated domains, which use 3D-structure information to explicitly to define domain boundaries, aligned blocks, and amend alignment details, and which aim to provide insights into how patterns of residue conservation and divergence in a family relate to functional properties.
- Pfam - a mirror of a recent Pfam-A database of curated seed alignments. Pfam version numbers do change with incremental updates. As with SMART, families describing very short motifs or peptides may be missing from the mirror. An HMM-based search engine is offered on the Pfam site.
- SMART - a mirror of a recent SMART set of domain alignments. Note that some SMART families may be missing from the mirror due to update delays or because they describe very short conserved peptides and/or motifs, which would be difficult to detect using the CD-Search service. You may want to try the HMM-based search service offered on the SMART site. Note also that some SMART domains are not mirrored in CD because they represent "superfamilies" encompassing several individual, but related, domains; the corresponding seed alignments may not be available from the source database in these cases. Note also that SMART version numbers do not change with incremental updates of the source database (and the mirrored CD-Search database).
- PRK - "PRK," short for Protein Clusters, is an NCBI collection of related protein sequences (clusters) consisting of Reference Sequence proteins encoded by complete prokaryotic and chloroplast plasmids and genomes. It includes both curated and non-curated (automatically generated) clusters.
- TIGRFAMs - a mirror of a recent TIGRFAMs set of domain alignments.
- COG - a mirror of the current COG database of orthologous protein families focusing on prokaryotes. Seed alignments have been generated by an automated process. An alternative search engine, "Cognitor", which runs protein-BLAST against a database of COG-assigned sequences, is offered on the COG site.
- KOG - a eukaryotic counterpart to the COG database. KOGs are not included in the CDD superset, but are searchable as a separate data set.
More information about each database is provided in the section on "Where does CDD content come from?" and the version number (as available) of each source database is provided in the CDD News page.
- Maximum number of hits: limits the size of the hit list produced by CD-Search. Typically, for average sized proteins, the number of expected domain-hits is small and the default setting of 500 should be more than sufficient.
- Expect Value (E-value): is a parameter that describes the number of hits one can "expect" to see by chance when searching a database of a particular size . The E-value setting can be modified to adjust the statistical significance threshold used for reporting matches against PSSMs in the database. False positive results should be very rare with the default setting of 0.01 (use a more conservative, i.e. lower, setting for more reliable results). Results with E-values in the range of 1 and above should be considered putative false positives. Additional information about E-value is available in the Glossary of the NCBI Handbook and in the BLAST help document .
Note that the E-values you get (for any given protein query--conserved domain hit pair) on the CD-Search web service might differ from those you get when using standalone RPS-BLAST on your local PC. A separate section of this document describes the differences between the web service and standalone program and provides a tip on how you can generate the same results in standalone RPS-BLAST as those produced by the web service.
- Composition-corrected scoring, which is employed by RPS-BLAST version 2.2.28 (March 19, 2013) and up, abolishes the need to mask out compositionally biased regions in query sequences. This option is on by default .
Note: In general, when composition-corrected scoring is on, the low complexity filter should be turned off. However, it is possible to have both options on at the same time (to filter false-positives that slip through the cracks of the composition-correction), or off at the same time (to find more distant relatives for compositionally biased queries), if desired.
- Low Complexity Filter: filters query sequences for compositionally biased regions. These regions are flagged as such and largely ignored during the search phase if filtering is turned ON (the default setting is OFF).
Note: In general, when the low complexity filter is turned on, the composition-corrected scoring should be turned off. However, it is possible to have both options on at the same time (to filter false-positives that slip through the cracks of the composition-correction), or off at the same time (to find more distant relatives for compositionally biased queries), if desired.If the low-complexity filter is turned on and compositially biased regions are detected, they are shown in the CD-Search output as cyan regions in the bar graphic that represents the query sequence, as illustrated below. More information about the low complexity filter is also available in the BLAST help document.
- If the low-complexity filter was ON for the search, the compositionally biased regions were NOT USED in the search against the domain database and are shown as SOLID cyan blocks. (As an example, open the default CD-Search results for P14780, GI 269849668, with filtering turned ON.) However, those regions may still overlap with or be included in a domain footprint and the pair-wise alignment generated by RPS-BLAST.

- If the low-complexity filter was turned OFF for the search, the compositionally biased regions were USED in the search and are shown as blocks OUTLINED in cyan. (As an example, open the CD-Search results for P14780, GI 269849668, with filtering turned OFF.) Please keep in mind, however, that compositionally biased regions can cause inaccurate annotation of the query sequence.

- If the low complexity filter DID NOT DETECT any compositionally biased regions in the query sequence, then it is displayed as a plain grey bar (with no cyan regions), as shown in the illustrations of the sample concise display and full display of CD-Search results.
- Force Live Search - Use this option if your query is a GI or accession number of a protein sequence already in the Entrez Protein database and you prefer to see live rather than precalculated CD-Search results. Note that precalculated searches use default parameters (options), while live searches allow you to change those parameters, if desired.
- Normally, CD-search will display precalculated search results for queries that contain a GI or accession number of a sequence already in the Entrez Protein database. This is because CD-Searches are done as part of the automated processing of the Entrez Protein database, and the stored search results are readily available. If that is true for your query, the " BLAST search parameters " information shown at the bottom of a Full Display of search results will say: Data Source: Precalculated Data, and will show the parameters (options) that are used by default.
- A Live search is done automatically IF: (a) your query is a FASTA formatted sequence, and the FASTA defline does not include a GI or accession number of a sequence record in Entrez protein, or (b) your query includes a GI or accession number but you selected a search database other than the default CDD or you changed any other parameters (options) from their default settings. If a Live Search was done, the " BLAST search parameters " information shown at the bottom of a Full Display of search results will say: Data Source: Live blast search, RID = XXNNNXXNNN. The RID is a "request ID" and will enable you to retrieve the results of that particular search for the next 36 hours. The display will also show the search parameters that were applied.
- Rescue Borderline Hits: This option allows you to see hits that have an E-value above the RPS-BLAST reporting threshold (anywhere between 0.01 and 1.0), and that are consistent with known domain architectures. A rescued hit is displayed with a dashed border, and its e-value is displayed in red, as shown in the illustrated example below. Additional details about rescued hits are provided in:
Derbyshire MK, Gonzales NR, Lu S, He J, Marchler GH, Wang Z, Marchler-Bauer A. Improving the consistency of domain annotation within the Conserved Domain Database. Database (Oxford) 2015 Mar 12; 2015. pii: bav012. doi: 10.1093/database/bav012. Print 2015. [PubMed PMID: 25767294] [Full Text at Oxford Journals] [Full Text in PubMedCentral]
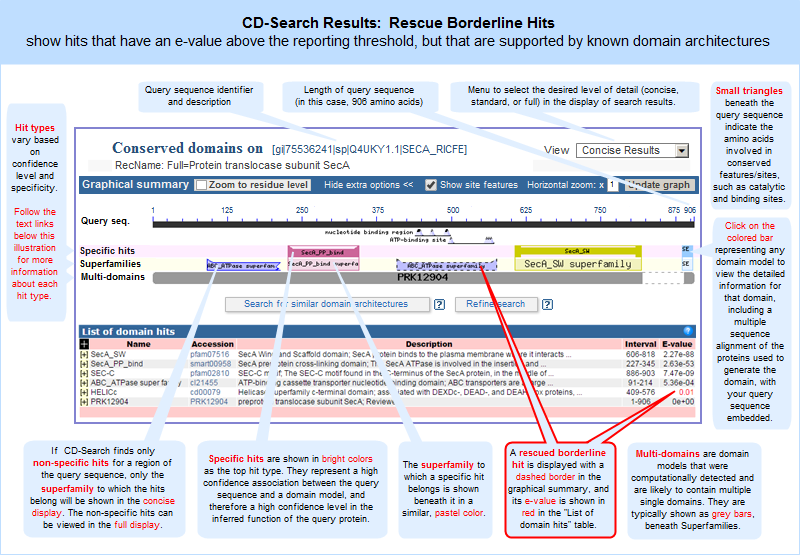 |
| The example above shows the search results, as of April 24, 2015, for protein GI 75536241 (Protein translocase subunit SecA). Hit types in the concise display can include specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models. Separate sections of this help document provide more information about the: (1) three levels of details available for viewing CD-Search results (concise display, standard display), full display); (2) display elements such as the protein classification, colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (3) display controls such as horizontal zoom and zoom to residue level; and (4) the options to search for similar domain architectures and refine search. Click anywhere on the illustration to open the current, interactive CD-Search: Concise results page for protein GI 75536241. (Note: The live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data and the refinement of algorithms to identify specific hits and superfamilies. However, the concepts shown in the illustration remain stable.) |
- Suppress Weak Overlapping Hits: This option suppresses hits that have an e-value close to the RPS-BLAST reporting threshold (in between 0.01 and 0.001) but overlap with stronger hits. A suppressed hit is displayed with a strikethrough, as shown in the illustrated example below. Additional details about suppressed hits are provided in:
Derbyshire MK, Gonzales NR, Lu S, He J, Marchler GH, Wang Z, Marchler-Bauer A. Improving the consistency of domain annotation within the Conserved Domain Database. Database (Oxford) 2015 Mar 12; 2015. pii: bav012. doi: 10.1093/database/bav012. Print 2015. [PubMed PMID: 25767294] [Full Text at Oxford Journals] [Full Text in PubMedCentral]
 |
| The example above shows the search results, as of April 24, 2015, for protein GI 29611879 (Alanine--tRNA ligase). Hit types in the concise display can include specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models. Separate sections of this help document provide more information about the: (1) three levels of details available for viewing CD-Search results (concise display, standard display), full display); (2) display elements such as the protein classification, colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (3) display controls such as horizontal zoom and zoom to residue level; and (4) the options to search for similar domain architectures and refine search. Click anywhere on the illustration to open the current, interactive CD-Search: Concise results page for protein GI 29611879. (Note: The live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data and the refinement of algorithms to identify specific hits and superfamilies. However, the concepts shown in the illustration remain stable.) |
- Result Mode: allows you to select the level of detail displayed in the search results: Concise mode (illustrated example) shows only the best scoring domain model, as available for each region on the query sequence. Standard mode (illustrated example) shows the best scoring domain model from each source database, for each region on the query sequence. Full mode (illustrated example) shows all hits for each region on the query sequence. Once you are viewing the search results, you can use the "View Full Result/View Concise Result" button in the upper right corner of the search results to toggle between the two views.
Retrieve a previous CD-Search result by entering its Request ID (RID):



A Request ID (RID) is assigned to a CD-Search if it was done as a live search. In such a case, the " BLAST search parameters " information shown at the bottom of a Full Display of search results will say: Data Source: Live BLAST search, RID = XXNNNXXNNN. The RID enables you to retrieve the results of that particular search for the next 36 hours by entering that number in the "Retrieve previous CD-Search result" section of the CD-Search page. (RIDs are not assigned to CD-Searches that use precalculated results.)
![]()
![]()
![]()
![]()
The CD-Search results page provides the following display options and information for the conserved domains that align to your query sequence:
- three levels of detail in CD-Search results displays: concise results | standard results | full results
- types of RPS-BLAST hits: specific | non-specific | superfamily | multidomain
- display elements: protein classification | domain colors/shapes | jagged edges (partial matches) | double-headed arrows (structural motifs) | small triangles (conserved features/sites) | compositionally biased regions
- display controls: horizontal zoom | zoom to residue level | refine search | search for similar domain architectures
- tabular list of domain hits
Three Levels of Detail in CD-Search Results Displays:



A pull-down "View" menu in the upper right corner of a search results page allows you to to select the desired view: Concise Results, Standard Results, or Full Results. This enables you to control the level of detail shown in both the Graphical Summary (shown in the illustrations below) and Tabular List of Domain Hits (not shown in the illustrations below for brevity, but available on the actual, interactive CD-Search results page for the example featured in the illustrations).
CD-Search results can include up to four hit types that represent various confidence levels (specific hits, non-specific hits) and scope (superfamilies, multi-domains) of domain hits. In the search result displays, hits are ranked by E-value, although NCBI-curated models are ranked ahead of other hits to the region if their E-value exceeds a threshold of 1e-05.
If the protein query sequence contains compositially biased regions, those will be detected by the low-complexity filter and shown in the graphical output as cyan regions in the grey bar graphic that represents the query sequence (illustrated example). The filter can be turned ON or OFF (default) when you submit a query by using the options on the search form.
Concise Results:



The Concise display is the default output for CD-Search results and shows only the best scoring domain model, as available for each region on the query sequence, in each of three hit types: specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models. In addition, small triangles (illustrated example) indicate the amino acids involved in conserved feaures/sites, such as catalytic and binding sites, when such annotations are available in a domain model.
If CD-Search finds both specific and non-specific hits for a region of a protein query sequence, only the highest ranking specific hit and its superfamily will be shown. If CD-Search finds only non-specific hits for a region of a protein query sequence, only the superfamily to which the hits belong will be shown, but not the non-specific hits themselves. The latter are provided only in the full display.
The top-scoring multi-domain model is shown in the concise display only if: (a) it meets or exceeds the specific hit threshold, OR (b) if it does not overlap with a specific hit or superfamily annotation and if ≥50% of the domain model's length aligns to the query protein sequence . If the top-scoring multi-domain model does not meet the 50% length threshold, it is shown on the concise display only if there is no specific hit or superfamily annotation on that query sequence region at all.
 |
| The example above shows the search results, as of October 22, 2014, for protein GI 157830769 (Cyclodextrin Glucanotransferase). Hit types in the concise display can include specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models. Separate sections of this help document provide more information about: (1) display elements such as the protein classification, colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (2) display controls such as horizontal zoom and zoom to residue level; and (3) the options to search for similar domain architectures and refine search. Click anywhere on the illustration to open the current, interactive CD-Search: Concise results page for protein GI 157830769. (Note: The live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data and the refinement of algorithms to identify specific hits and superfamilies. However, the concepts shown in the illustration remain stable.) |
Standard Results:



The Standard result lists the best scoring domain model from each source database, as available for each region on the query sequence . In some cases, two NCBI-curated models might be shown for a given region of a protein, if the immediate parent of the highest ranking NCBI-curated conserved domain model is also in the search results. (A separate section of this document provides more information about domain family hierarchies.) The top-scoring multi-domain model from each source database is also shown.
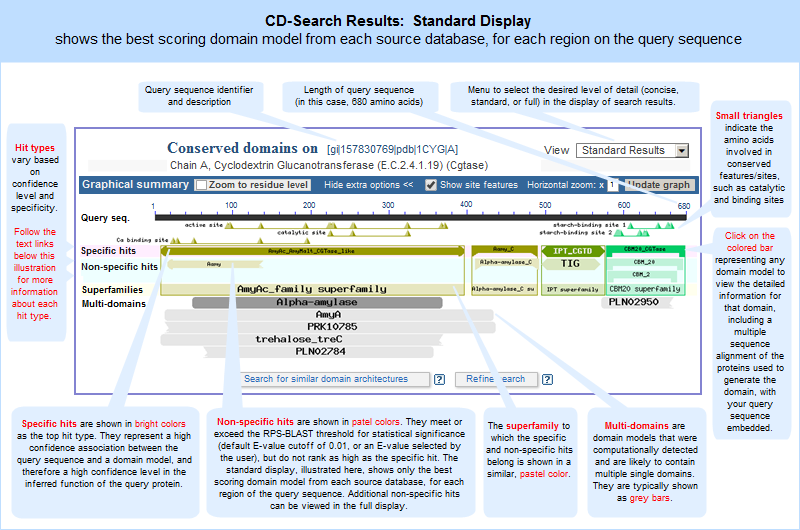 |
| The example above shows the standard search results, as of October 22, 2014, for protein GI 157830769 (Cyclodextrin Glucanotransferase). Hit types in the standard display can include specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models. Separate sections of this help document provide more information about: (1) display elements such as the protein classification, colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (2) display controls such as horizontal zoom and zoom to residue level; and (3) the options to search for similar domain architectures and refine search. Click anywhere on the illustration to open the current, interactive CD-Search: Standard results page for protein GI 157830769. (Note: The live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data and the refinement of algorithms to identify specific hits and superfamilies. However, the concepts shown in the illustration remain stable.) |
Full Results:



The Full display shows all domain models, as available for each region on the query sequence, that meet or exceed the RPS-BLAST threshold for statistical significance (i.e., the E-value cutoff). The hit types can include specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Hits are ranked by E-value, although NCBI-curated models are ranked ahead of other hits to the region if their E-value exceeds a threshold of 1e-05. In addition, small triangles indicate the amino acids involved in conserved feaures/sites, such as catalytic and binding sites, when such annotations are available in a domain model.
The bottom of the Full Display (not shown in the image below but viewable by clicking on that image to open the actual, interactive CD-Search results page) also provides a summary of BLAST search parameters, which includes information such as the database which you searched against, whether the low complexity filter was used, the expect value (E-value) threshold, the BLAST software version number, and whether RPS-BLAST did a live search or retrieved precalculated search results. If a live search was done, the BLAST Request ID (RID) is also shown in the "BLAST search parameters" section and allows you to retrieve the search results by RID anytime within 36 hours following the search, without having to re-execute it.
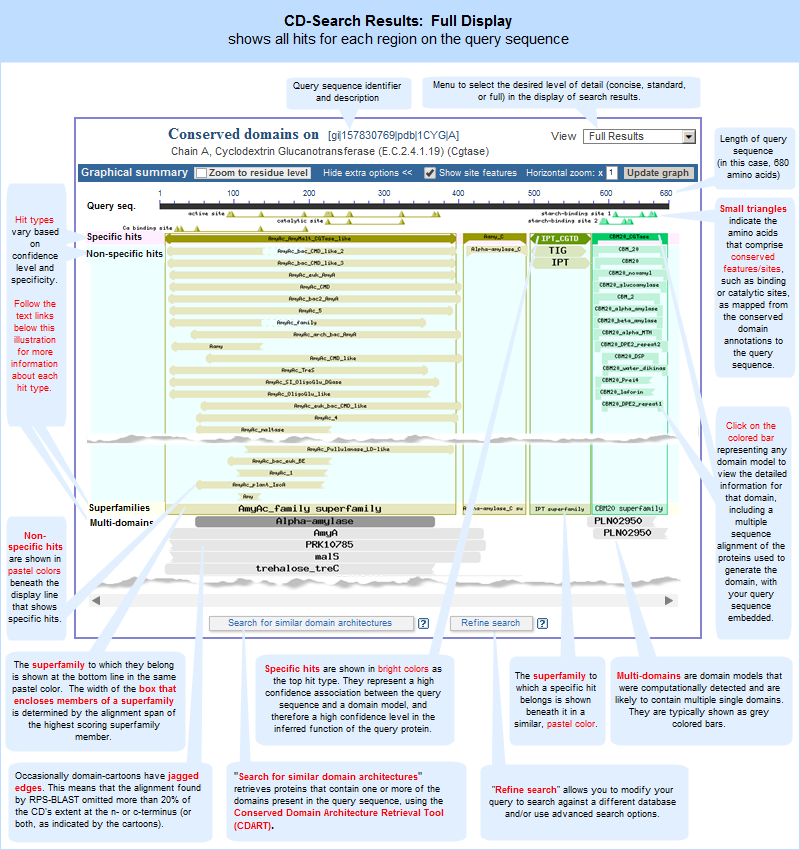 |
| The example above shows the search results, as of October 22, 2014, for protein GI 157830769 (Cyclodextrin Glucanotransferase). Hit types in the full display can include specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Separate sections of this help document provide more information about: (1) display elements such as the colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (2) display controls such as horizontal zoom and zoom to residue level; and (3) the options to search for similar domain architectures and refine search. Click anywhere on the illustration to open the current, interactive CD-Search: Full results page for protein GI 157830769. (Note: The live web page may look different from the illustration shown here, because the Conserved Domain Database continues to evolve with the addition of new data and the refinement of algorithms to identify specific hits and superfamilies. However, the concepts shown in the illustration remain stable.) |
Types of RPS-BLAST hits:



CD-Search results can include hit types that represent various confidence levels (specific hits, non-specific hits) and domain model scope (superfamilies, multi-domains). They can be seen in both the Concise display and Full display, except for non-specific hits, which are shown only in the Full Display.
- Specific hit is the top-ranking RPS-BLAST hit (compared to other hits in overlapping intervals) that meets or exceeds a domain-specific E-value threshold (details and illustration) . It represents a very high confidence that the query sequence belongs to the same protein family as the sequences used to create the domain model, and therefore a high confidence level for the inferred function of the protein query sequence.
- Non-specific hits meet or exceed the RPS-BLAST threshold for statistical significance (default E-value cutoff of 0.01, or an E-value selected by the user with advanced search options). (NOTE: Non-specific hits are shown only in the full display (illustration) of search results. In contrast, the concise display (illustration) shows only the superfamily to which the top-scoring non-specific hit for a given sequence region belongs.)
- Superfamily is the domain cluster to which the specific and/or non-specific hits belong. This is a set of conserved domain models that generate overlapping annotation on the same protein sequences and are assumed to represent evolutionarily related domains. (See additional details, including information about clustering methodology, under "What is a superfamily?") In the Concise Display, if a region of the query sequence has only non-specific hits to domain models from a given superfamily, only the superfamily footprint will be displayed -- not the individual superfamily members to which the query sequence had non-specific hits. To see the latter, view the Full Display of search results. In that display, the width of the box that encloses superfamily members is determined by the alignment span of the highest scoring superfamily member.
- Multi-domains are domain models that were computationally detected and are likely to contain multiple single domains. They are typically shown as grey-colored bars. (Examples are shown in the concise display and full display illustrations.)
Display Elements:



A number of display elements are used to graphically convey conserved domain annotations on the query sequence. Those elements are used in all three views of search results: Concise Results, Standard Results, or Full Results. The display elements, described below, include:
The Tabular List of Domain Hits, which appears beneath the graphical summary of search results, provides additional details about, and viewing options for, each conserved domain model that has been mapped to your query sequence. Each domain model's accession number links to the corresponding record in the Conserved Domain Database, where the model and its member sequences can be launched in free software such as the Cn3D structure viewing program, or the CDTree alignment viewer/editor.
- protein classification
- domain colors/shapes
- jagged edges (partial matches)
- double-headed arrows (structural motifs)
- small triangles (conserved features/sites)
- compositionally biased regions
Protein Classification:



- How is the protein classification determined?
A protein classification is shown in the CD-Search results, when possible, and provides a functional characterization of the conserved domain architecture found in the protein query (illustrated example). The protein classification section appears in CD-Search results only if we have a curated label for the protein family to which the query sequence belongs.
A domain architecture is defined as the sequential order of conserved domains in a protein, and the architectures are computationally identified by the Conserved Domain Architecture Retrieval Tool (CDART).
A tool called
SPARCLE (Subfamily Protein Architecture Labeling Engine) is used to label the proteins that contain a given architecture.Each conserved domain architecture can be assigned a unique, functional name based on the composition of the architecture. The names are assigned either manually, through a curation process, or computationally, by an autoname algorithm or a namedbydomain algorithm.
The domains used for architectures may include ancient superfamilies (like ATPase) or much more recently evolved protein subfamilies (like RAS).
In the case of curated architectures, the functional characterization of each architecture is written by Conserved Domain Database Curators, based on a review of the publications associated with the proteins that contain the domain architecture.
To given an example of proteins that have similar function but different domain architectures:
There are several types of architectures:
- DNA gyrase B (NP_387887), an antibiotic target, has a conserved domain architecture that includes a histidine kinase-like ATPase domain, a transducer domain, a topoisomerase-primase domain, followed by a type II topoisomerase carboxy domain .
- In contrast, enzymes of similar function, such as topoisomerase IV (Q45066), have a different domain architecture.
Note: in each of the examples above, the default graphic that appears when you click on the architecture link depicts the full length protein model; click on the option to "View: Full Results" link in the upper right hand corner of the display to see the individual conserved domains that compose the full length protein model.
Also, in each example, you can follow the "domain architecture ID xxxxxxx" link that appears in the "Protein Classification" section of the display to open the corresponding SPARCLE record. The SPARCLE record, in turn, lists the evidence that was used to name the architecture and contains links to other protein sequences that have the same architecture.
Separate sections of this help document provide additional information about domain family hierarchies, and the hit types you see in CD-Search results, such as specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other.
- superfamily architectures - domain architectures consisting solely of superfamilies.
- subfamily architectures - domain architectures that mix superfamilies and subfamilies (i.e., conserved domain models that get a specific hit to the protein query sequence).
Note: It is also possible for a domain architecture to consist of a single conserved domain footprint .
The SPARCLE Help document provides additional information about that resource, including an overview, examples of how SPARCLE can be used to learn more about proteins, allowable input, a description of the search output and the contents of a SPARCLE record, and details about the data processing pipeline.
Domain colors:



- What is the significance of the domain colors/shapes?
Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other. (Because the number of superfamilies exceeds the number of different color/shape combinations, some of the superfamilies share the same display style. It should be rare, though, to encounter such pairs in the same domain architecture.)
The color/shape combination remains stable for a given superfamily cluster ID, and is consistent across all NCBI tools that show conserved domain footprints on protein sequences (i.e., CD-Search, Batch CD-Search, and CDART).
If the cluster ID for a superfamily changes (due to significant changes in the superfamily composition), the color/shape combination may also change.
"Specific hits" are shown as bright colors, with full saturation. The "superfamily" to which a "specific hit" belongs is shown as a pastel color, with lower saturation.
If there are no specific hits to a region of a protein query sequence, then the "Concise display" will show only the superfamily. Regardless whether there are specific hits or not, "Standard display" will show the superfamily plus highest ranking hit from each source database, and the "Full display" will show the all the domain models in each hit type (specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models).

Jagged Edges:



- What do domain cartoons with jagged edges mean? Occasionally domain-cartoons have jagged edges (illustrated example). This means that the alignment found by RPS-BLAST omitted more than 20% of the CD's extent at the n- or c-terminus (or both, as indicated by the cartoons). This feature may give hints towards truncated query sequences, false-positive hits, or unusual domain architectures involving long insertions. The exact percentage of the CD's extent used in the alignment is listed in detail in the pairwise alignment section.
Double-headed Arrows (structural motifs)



- What do the double-headed arrows mean in the Graphical Summary?
Double-headed arrows appear on a CD-Search results graphical summary only if the query protein contains structural motifs. Structural motifs are regions in proteins and protein domains that are too small to be modelled as individual evolutionarily conserved domains and too extensive to be characterized as
conserved features/sites. They play a structural and/or functional role that CDD curators chose to document, as their presence contributes to functional annotation and/or protein classification. Structural motifs are particularly useful in annotating the locations of specific repeats. Examples are blades in beta-propeller structures ("closed solenoid proteins"), super-helical repeats such as Armadillo and HEAT ("open solenoid proteins"), various zinc fingers, various calcium-binding motifs, coiled coils, or transmembrane segments.The structural motifs cannot be modeled as evolutionarily conserved domains because the properties of the PSSMs as search models require a minimum length to be effective (exacerbated by the fact that many of the structural motif regions have compositional bias), and because the evolutionary history of most of these structural motifs is not clear enough to enable a representation of that history.
 |
| The example above shows the search results, as of October 22, 2014, for protein GI 355339453 (neuraminidase from Influenza A virus). The double-headed arrows that appear beneath the query sequence represent structural motifs. Click anywhere on the illustration to open the current, interactive CD-Search results page for protein GI 355339453. The CD-Search results display can include various hit types, such as specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other. Separate setions of this help document provide more information about: (1) the three levels of detail available in CD-Search search results displays: concise results, standard results, and full results; (2) display elements such as the colors/shapes used for the domain cartoons, the small triangles that represent conserved features/sites, the double-headed arrows that represent structural motifs; (3 display controls such as horizontal zoom and zoom to residue level; and (4) the options to search for similar domain architectures and refine search. |
Small Triangles



- What do the small triangles mean in the Graphical Summary?
The small triangles beneath the query protein on a CD-Search results page indicate the residues that comprise conserved features/sites, such as binding or catalytic sites, as mapped from the conserved domain annotations to the query sequence. An illustrated example is below.
The triangles appear if a region of the query protein sequence either:
- gets a specific hit to an NCBI-curated domain domain model on which conserved features/sites have been annotated. In such a case, the conserved features/sites that have been annotated on the domain model will be mapped to the query sequence.
OR
- gets a non-specific hit to an NCBI-curated domain model that belongs to a superfamily whose representative has conserved/feature site annotations. In such a case, the conserved features/sites from the superfamily representative will be mapped to the query sequence. (Note that the non-specific hit will not appear on the concise display of the CD-Search results -- only the site annotations will appear there. View the full display to see both the triangles and the hit.)
The triangles are shown in the same color as the domain on which they have been annotated.Click on the triangles to view details about the feature, including a multiple sequence alignment of your query sequence and the protein sequences used to curate the domain model, where hash marks (#) above the aligned sequences
(illustration) show the location of the conserved feature residues. A thumbnail image, if present, provides an approximate view of the feature's location in three dimensions and options for interactive 3D structure viewing. Conserved features/sites, if present, are shown by default in the graphical display. If desired, they can be hidden by clicking on show options in the graphical summary header bar, then deactivating the show site features checkbox and pressing the update button.
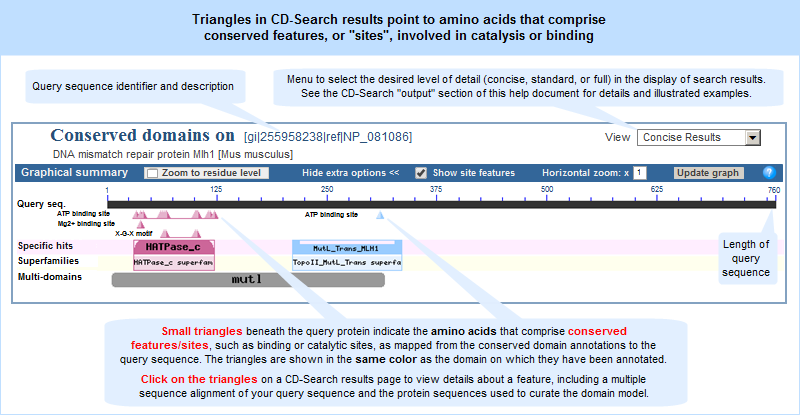 |
| The example above shows the search results, as of October 22, 2014, for protein GI 255958238 (mouse DNA mismatch repair protein Mlh1). Click anywhere on the graphic to view the actual, interactive CD-Search results page. The hit types in the full display can include specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other. If conserved features/sites are present, triangles are shown all three types of search results displays (concise results, standard results, and full results). |
Compositionally Biased Regions:



- On the CD-Search results page, what do the cyan regions mean in the bar graphic that represents the query sequence?
These represent compositially biased regions detected in the query sequence by the low-complexity filter.
- If the low-complexity filter was ON for the search, the compositionally biased regions were NOT USED in the search against the domain database and are shown as SOLID cyan blocks. (As an example, open the default CD-Search results for P14780, GI 269849668, with filtering turned ON.) However, those regions may still overlap with or be included in a domain footprint and the pair-wise alignment generated by RPS-BLAST.

- If the low-complexity filter was turned OFF (default) for the search, the compositionally biased regions were USED in the search and are shown as blocks OUTLINED in cyan. (As an example, open the CD-Search results for P14780, GI 269849668, with filtering turned OFF.) Although compositionally biased regions can cause inaccurate annotation of the query sequence, their effect is ameliorated to a great extent by composition-corrected scoring, which is turned on by default.

- If the low complexity filter DID NOT DETECT any compositionally biased regions in the query sequence, then it is displayed as a plain grey bar (with no cyan regions), as shown in the illustrations of the sample concise display and full display of CD-Search results.
Display Controls:



The CD-Search results display can be customized with the following controls:
The Tabular List of Domain Hits, which appears beneath the graphical summary of search results, provides additional details about, and viewing options for, each conserved domain model that has been mapped to your query sequence. Each domain model's accession number links to the corresponding record in the Conserved Domain Database, where the model and its member sequences can be launched in free software such as the Cn3D structure viewing program, or the CDTree alignment viewer/editor.
- horizontal zoom
- zoom to residue level
- refine search
- search for similar domain architectures
Horizontal Zoom



- If a query sequence is very long and contains many domains (e.g., human titin isoform N2-B, gi 291045223), the details of the graphical summary might be difficult to read. In that case, you can click on show options in the graphical summary header bar, enter the desired magnification level in the horizontal zoom box, and press the update button to refresh the display.
There is no specific maximum value that can be entered in the horizontal zoom box. Rather, the limit is determined by the pixel width of the graphic image displayed.
- If the zoom value you enter is too large, the system will display the message: "invalid zoom factor". In that case, enter a smaller zoom value.
- There might be other cases in which the zoom value is acceptable but it takes some time to generate the display. In such cases, you might get an option to stop script or continue. Choose the latter if you would like the process of generating the enlarged graphic display to continue.
- Zoom to Residue Level




- This option displays the amino acids ("residues") in the query sequence. It also highlights the amino acids in the query that are mapped to conserved features/sites , which are denoted by small triangles in the graphical summary. As an example of the "zoom to residue level" view, see the human regulator of G-protein signaling 12 isoform 2.
- When you activate the "zoom to residue level" setting, the "horizontal zoom" text box (which is visible when you press "show extra options") will still retain the zoom value that was used before the "zoom to residue level" option was activated. This makes it possible to easily toggle between the residue level view and the previous zoom level.
Note: The "show extra options/horizontal zoom" text box will generally contain the default value of 1, unless you viewed a different magnification before zooming in to the residue level. The actual horizontal zoom level that is applied by the CD-Search program when the "zoom to residue level" option is checked varies based on length of sequence and is determined automatically by the program.
- Note about display limits: If a protein sequence is very long, the individual amino acids might not be visible when the "zoom to residue level" option is checked. This is because images wider than approximately 35000 pixels cannot be displayed correctly in browsers. Therefore, the "zoom to residue level" option limits the display to 35000 pixels wide. If the query sequence is very long (as an example, see human titin isoform N2-B, 26,926 amino acids long), the program will still draw the residues, but they will be squeezed together and will not be easily readable as individual letters.
Refine Search



- The Refine Search button on a CD-Search results page allows you to modify your query to search against a different database and/or use advanced search options.
Search for Similar Domain Architectures



- The Search for Similar Domain Architectures button on a CD-Search results page retrieves proteins that contain one or more of the domains present in the query sequence, using the Conserved Domain Architecture Retrieval Tool, "CDART" ( illustrated example).
Tabular List of Domain Hits



- Beneath the CD-Search results graphical summary is a Tabular List of Domain Hits. This table appears in all three views: Concise Display, Standard Display, and Full Display.
When you mouse over any conserved domain footprint in the graphical summary, the corresponding CD accession number and description will be highlighted in the tabular list of domain hits.
If a domain model aligns to more than one region of the query sequence, it will be listed multiple times in the tabular list of domain hits. This is true because the alignment coordinates and score of the domain model vary among different regions of the query sequence, and each hit is reported separately.
Click on the [+] to the left of the CD accession to see a pairwise alignment of your query sequence and the consensus sequence for the domain model. (Residues that are identical between your query sequence and the consensus sequence are shown in red .) Click on the CD accession number to view the domain model's summary record in the Conserved Domain Database (CDD).
If you'd like to see your
query sequence embedded in the domain model's multiple sequence alignment , click on the domain footprint for any specific hit, non-specific hit, or multidomain of interest in the graphical portion of the concise or full CD-search results page. That will open the Entrez CDD record for the domain model, with your query sequence embedded in the model's multiple sequence alignment. In that view, you can change the color bits setting to increase or decrease the threshhold that determines which columns of the alignment are displayed in red . (Note: Superfamily records do not include a multiple sequence alignment display, so if you click on the footprint of any superfamily, you will see a CDD summary page that provides general information about the superfamily and lists the domain models that belong to it. Only the individual domain models will have multiple sequence alignments, and you must click on the footprints of those models in the graphical summary of a CD-Search results page (not on the superfamily's CDD summary page) in order to see your query sequence embedded in the alignment.)The tablular list of domain hits also provides a link from each domain model's accession number to the corresponding record in the Conserved Domain Database, where the model and its member sequences can be launched in free software such as the Cn3D structure viewing program, or the CDTree alignment viewer/editor.
![]()
![]()
![]()
![]()
A specific hit is a high confidence association between a protein query sequence and a conserved domain, resulting in a high confidence level for the inferred function of the protein query sequence. It is one of four types of RPS-BLAST Hits. (See illustrations of CD-Search results concise display and full display for examples.)
In order to be considered a specific hit, an alignment of a domain model to a query protein sequence must meet two criteria:
- The domain model must be either: (1) the top-ranked (best E-value) NCBI-Curated domain, or (2) the top-ranked domain model from an external source, if there is no NCBI-curated domain that meets all the criteria for a specific hit.
If domain models from both the NCBI-curated data set and external sources meet a domain-specific threshold, the NCBI-Curated domain domain will be listed preferentially as the specific hit because it has been annotated with fine-grained evolutionary relationships, conserved sequence blocks, specific functions, and conserved features/sites based on careful review of sequence data, 3D structures, and literature.
However, if no NCBI-curated domain meets the criteria for a specific hit, then the top-ranked domain model from an external source will be shown in the CD-Search results concise display if it meets all the criteria for a specific hit.- The E-value of the RPS-BLAST hit must be equal to or lower than a domain-specific threshold E-value.
The domain-specific threshold is the weakest E-value obtained when each of the protein sequences used to curate a domain are RPS-BLAST'ed against that domain's Position-Specific Scoring Matrix (PSSM). In other words, the threshold is the weakest E-value among self-hits of a domain�s member protein sequences to the resulting domain model. The illustration below provides an example, showing the domain-specific threshold for cd03683, ClC-1-like chloride channel proteins. Domain-specific threshold scores are displayed (in the form of bit score) in the statistics box of a domain model's CD-summary page.
| ADDITIONAL DETAILS: If multiple NCBI-Curated domain models align to a given interval on a query protein sequence and pass both of the criteria above, then the highest-scoring model is the specific hit and the other models are listed as non-specific hits. The highest scoring model is in general the one with the best E-value, but if two or more models have the same E-value, then their bit score is used to break the tie. For example, the CD-Search results for protein sequence NP_229631 show several NCBI-curated domains aligned to the same region of the query. The top-ranked NCBI-curated domains are cd05297 (GH4_alpha_glucosidase_galactosidase) and cd05197 (GH4_glycoside_hydrolases), both of which have an E-value of 2e-169 (as of 08 March 2010). However, the bit score for the hit to cd05297 (590.69) is higher than the bit score for cd05197 (590.65), so cd05297 is displayed in the CD-Search results as the specific hit and cd05197 is displayed as a non-specific hit. In the unlikely event that bit score is insufficient to break the tie, only one hit is randomly chosen to be a specific hit. (Note: The bit score of a CD-Search hit to a domain model can be seen by clicking on the plus (+) to the left of its accession number in the tabular "List of Domain Hits" on the CD-Search results page. Additionally, the domain-specific threshold bit score for an NCBI-curated domain is displayed in the statistics box of the domain model's CD summary page.) In contrast, some protein query sequences can have several hits to NCBI-curated domains, and none of them will show up as a specific hit. That is true in the CD_Search results for protein sequence NP_486772 (as of 08 March 2010). In that case, cd01662 (Ubiquinol oxidase I) is the top-ranked (best E-value) NCBI-curated domain; however, it is not shown as a specific hit because the bit score of that hit does not meet or exceed the domain-specific threshold. The hits to two other NCBI-curated domains, cd01663 (Cyt_c_Oxidase_I) and cd00919 (Heme_Cu_Oxidase_I), have bit scores that meet or exceed the domain-specific thresholds for those models, but they are not listed as specific-hits because neither one of them is the top-ranked (i.e., best E-value) NCBI-curated domain. TO SUMMARIZE: In order to be a specific hit, a domain model must: (a) be the top-ranked domain model *AND* (b) have a bit score that meets or exceeds the domain-specific threshold score. Combining the two criteria was found to reduce the number of false positive calls. |
If a specific hit IS found on a protein sequence, then:
- There is a high confidence level that the query protein sequence is a member of the protein family represented by the domain model and has the specific function annotated on that domain.
- If the query sequence resides in the Entrez Protein database, the inferred function is annotated as "region" on the protein sequence record, showing the name of the high-scoring domain model and its base span. If the specific hit is to an NCBI-curated domain model that includes conserved features (residues involved in catalysis or binding), those are annotated on the protein sequence record as "sites." If the specific hit is to a domain model from an external source, and the model belongs to a superfamily whose representative is an NCBI-curated domain that has such annotations, then the conserved features/sites that have been annotated on the superfamily representative will be mapped to the query sequence.
If a specific hit IS NOT found on a query protein sequence, but the protein has an otherwise statistically significant hit (E-value cutoff of 0.01) to any domain model in CDD, the domain model is regarded as a non-specific hit. In that case:
- The general function of the domain superfamily can be inferred for the query protein sequence, but the specific function is less certain.
- If the query protein sequence resides in the Entrez Protein database, the name and general function of the domain superfamily is annotated in the protein sequence record (as a "region"). The name and function text is derived from the domain model which has been selected as the superfamily representative. Conserved features ("sites") are also annotated on the protein sequence record if the superfamily representative is an NCBI-curated domain that has such annotations.
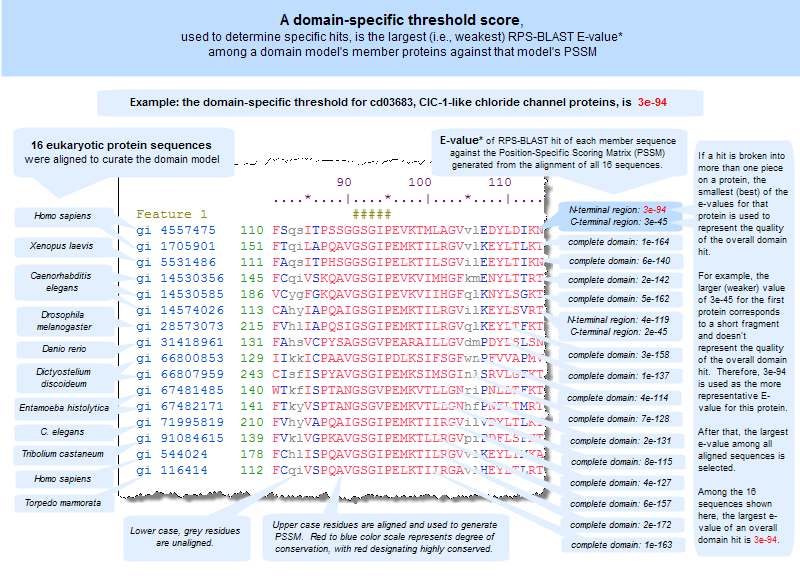 * Domain-specific threshold scores are displayed in the statistics box of a domain model's CD summary page. In the actual calculation of domain-specific thresholds, bit scores are used rather than E-values. (A bit score is defined in the NCBI Handbook glossary and Field Guide glossary.)
* Domain-specific threshold scores are displayed in the statistics box of a domain model's CD summary page. In the actual calculation of domain-specific thresholds, bit scores are used rather than E-values. (A bit score is defined in the NCBI Handbook glossary and Field Guide glossary.)
NOTE: The image above reflects the cd03683 domain alignment as of April 20, 2008 . The scientific community's understanding of molecular data continues to evolve as research progresses, and as new as well as updated sequence data are regularly deposited into the databases. If a member sequence used in a domain alignment is later superceded by an updated version, the new sequence data and gi number will replace the old one during review/update cycles of curated domains. Some revisions to sequence data, such as upstream or downstream extensions, do not affect the domain model, but the gi number and amino acid span will change to reflect the updated sequence record.
![]()
![]()
![]()
![]()
When you click on the cartoon (colored bar representing a domain footprint) in the graphical display on the CD-search results page, an alignment view will be opened, which adds the query sequence to the multiple CD-alignment. It is possible to modify the number and type of sequences shown, as described in the help document section on CDD Record : multiple sequence alignment displays .
![]()
![]()
![]()
![]()
If you display an alignment view that includes a query sequence, you can also view the same alignment in the Cn3D program by pressing the Structure View button. (Cn3D installation takes only a couple of minutes and a tutorial describes the program's features and functions. The program must be installed in order for the Structure View button to work.)If a protein sequence from a 3D structure is included among the sequences used to curate a domain model, Cn3D will show the 3D structure as well. If the domain model includes sequences from more than one 3D structure, all of the structures will be displayed, superimposed upon each other, and their sequences will be displayed in the multiple sequence alignment.
Cn3D offers column-specific coloring by sequence conservation when invoked with multiple alignment views. This is a convenient feature to study sequence conservation within a CD-alignment and to find out how well the aligned query fits the existing patterns of conservation and variability.
![]()
![]()
![]()
![]()
CD-search requests are submitted to the BLAST servers immediately. A typical search should take a few seconds only, depending on the size of the search database chosen, the length of the query sequence, and the load on the servers. Click here to test response time with a typical query.
CD-Search requests can also be sent to the BLAST Queuing system (this happens by default for searches launched in parallel with protein BLAST requests), use the optional button at the bottom of the CD-Search page. Requests sent to the query will take longer, but the results can be retrieved at a later time using the RID ("Request ID"), without having to re-calculate the search. A form at the bottom of the CD-Search page can be used to retrieve earlier search results by RID.
![]()
![]()
![]()
![]()
When CD-search is run as an integral part of protein-BLAST search requests, the jobs are put in the BLAST queue and may take a little longer to complete (depending on the system load and length of query sequence). Queued CD-search will try to retrieve the finished results every few seconds until they are available. You may also store the request-id (RID) and retrieve results later here.
Yes, you can run RPS-BLAST locally. A standalone version of RPS-BLAST is packaged in with the BLAST executables available on the NCBI FTP site, and is also available as part of the NCBI toolkit distribution (see ftp://ftp.ncbi.nih.gov/toolbox).Separate directories on the FTP site provide documents that describe each of the BLAST applications, including documents for RPS-BLAST and a Formatrpsdb application that can be used to build search databases that are properly formatted for use with RPS-BLAST.
Pre-formatted search databases, which have already been processed by Formatrpsdb, are available on the CDD FTP site. A README file on the CDD FTP site also provides more details about customizing search databases.
![]()
![]()
![]()
![]()
There are several differences between the CD-Search web service and standalone RPS-BLAST, as distributed by NCBI and used with search databases as distributed by the CDD group.The web server is optimized for the most common use of the CDD resource, which is to annotate protein sequences with clearly identified and well understood protein domains, and is also optimized for speed in order to accomodate a high volume of searches.
As part of the optimization, we use some different statistical parameters for the web service than for the standalone RPS-BLAST application. Specifically, we use a constant, assumed search "database size" setting on the web server for calculating E-values. This means that the actual size of the search database can change (we are adding new models every few weeks), but the E-value computed for any individual GI -- PSSM match will remain constant. This approach: (a) ensures that pre-calculated residues are not dependent on the actual size of the model collection (which is redundant and mostly grows by increasing that redundancy); (b) facilitates incremental updates of
pre-computed sequence annotation with conserved domains; and (c) is used for the creation of protein-CDD links.In contrast, standalone RPS-BLAST does not employ the constant, assumed database size parameter. So when you use a search set downloaded from the CDD FTP site, the database size might be different than the one used by the CD-Search web service, and the same hit of your query protein to a model will receive a different E-value in the standalone result. For example, if the size of the FTP'ed database is smaller than what the CD-Search web service assumes in its database size parameter, the same hit of your query protein to a model will receive a lower
E-value in the standalone. Conversely, if the size of the FTP'ed database is larger than what the CD-Search web service assumes in its database size parameter, the same hit of your query protein to a conserved domain model will receive a higher E-value in the standalone.If you want standalone RPS-BLAST to use the same database size parameter that is used for the web server (and thereby reproduce the same E-values with standalone RPS-BLAST that are generated by the web service), you can do that by creating an "alias" file on your local computer and placing it in the same directory as the standalone RPS-BLAST executable. The file can have a name such as "mycdd.pal" and can have contents such as the following (where lines starting with "#" are comments):
# # RPSBLAST alias file # TITLE mycdd # DBLIST ./Cdd # STATS_TOTLEN 5000000 STATS_NSEQ 21000This will now let you search against the database named "Cdd" using the two search set size parameters as specified, e.g.:~$ rpsblast -i rpstest.tfa -d mycdd -F T -e 0.01 -m 9 # RPSBLAST 2.2.26 [Sep-21-2011] # Query: gi|156356500|ref|XP_001623960.1| predicted protein [Nematostella vectensis] # Database: mycdd # Fields: Query id, Subject id, % identity, alignment length, mismatches, gap openings, q. start, q. end, s. start, s. end, e-value, bit score gi|156356500|ref|XP_001623960.1| gnl|CDD|197660 31.91 47 29 2 432 475 4 50 7e-04 36.9 gi|156356500|ref|XP_001623960.1| gnl|CDD|197660 31.48 54 31 3 493 545 6 54 8e-04 36.5 gi|156356500|ref|XP_001623960.1| gnl|CDD|197660 33.33 42 27 1 312 352 2 43 0.003 35.3 gi|156356500|ref|XP_001623960.1| gnl|CDD|119391 23.53 51 34 2 493 542 1 47 8e-04 36.4 gi|156356500|ref|XP_001623960.1| gnl|CDD|119391 21.57 51 35 2 375 424 1 47 0.003 34.5 gi|156356500|ref|XP_001623960.1| gnl|CDD|177721 24.47 94 56 3 463 541 18 111 0.005 38.6In addition to the different statistical parameters, the CD-Search web service does not filter out compositionally biased regions in the query sequence by default. It uses composition-corrected scoring to mitigate the effects of compositional bias. In contrast, standalone RPS-BLAST filters out compositionally biased segments and does not employ composition-corrected scoring. In the current RPS-BLAST version 2.2.29 (as of Feb. 2014), you can set parameters to replicate CD-Search settings by specifying "-comp_based_stats 1" and "-seg no" on the command line. If those options are not specified, standalone RPS-BLAST may retrieve somewhat different results. Finally, some advanced options in standalone RPS-BLAST are not available in the web service, such as the ability to use a single-hit/two-pass mode in order to detect more distant homologous relationships . Users who select such options in the standalone version may get different search results with the web service.
| What is batch CD-Search? | input | output | scripted data downloads | references |
![]()
![]()
![]()
![]()
Batch CD-Search serves as both a web application and a script interface for a conserved domain search on multiple protein sequences , accepting up to 4,000 proteins in a single job. It uses RPS-BLAST to compare a batch of query protein sequences against conserved domain models that have been collected from a number of source databases, then allows you to view a graphical display of the concise or full search result for any individual protein from your input list, or to download the results for the complete set of proteins. (To search for conserved domains on a single protein sequence, the original CD-Search service continues to be available; see the CD-Search help for more details.)
![]()
![]()
![]()
![]()
Protein sequences only | protein unique identifiers (UIDs) or protein sequences | maximum input
Adjust search options: search mode, database selection, expect value, composition-corrected scoring, low complexity filter, maximum hits, include retired sequences
Optional job title | e-mail notification when job is done | retrieve previous search result
- Protein sequences only




- Batch CD-Search accepts only protein sequences . The maximal number of queries per request is 4000, as noted under maximimum input, below. (Standard CD-Search, which is used to input individual queries, can accept either protein or nucleotide sequences.)
- A list of query proteins can be entered directly (typed or copied/pasted) into the text box on the Batch CD-Search web page or uploaded as a text file. (A separate section of this document describes scripted data downloads.)
- The query proteins can be represented as a list of sequence identifiers or as sequence data, separated by line breaks, as described below.
- Each job receives a randomly generated, unique Search ID.
- Note: If multiple query proteins are inadvertently entered on the regular CD-Search page, your query will be automatically redirected to the Batch CD-Search tool. If there are no line breaks between the query proteins, however, an error message will be displayed and no redirect will occur. Also note that Batch CD-search does not accept nucleotide sequences; nucleotide sequence queries, however, can be submitted individually to the standard CD-Search tool.
- Protein unique identifiers (UIDs)




- GIs or Accessions -- protein query sequences can be entered into Batch CD-Search as GI numbers (e.g., 230702), accession numbers (e.g., 2RMU2), or vertical line delimited NCBI style identifiers (e.g., gi|230702). If the protein sequences are molecules from PDB records, their identifiers should be entered as PDB accessions combined with chain codes. For example, the identifiers for the three protein chains from the three-dimensional structure 1TUP (Tumor Suppressor P53 Complexed With Dna) would be entered as 1TUPA, 1TUPB, 1TUPC.
Each sequence identifier, regardless of format, should occupy a single line (end with a new line character). Multiple identifiers in one line will be treated as one and will result in an "invalid identifier" error, and will therefore be ignored by Batch CD-Search.
- UID validation -- After you input a UID list into Batch CD-Search, the tool checks your list to ensure all the identifiers are valid. Invalid identifiers will be ignored, and the program will return results only for the valid identifiers.
In order to be considered VALID, a GI or accession must be present in (1) the live Entrez Protein database (these are considered CURRENT identifiers) OR (2) in the backend, archival database
, which contains CURRENT as well as NOT CURRENT (i.e., deprecated or preliminary) identifiers.Note that NOT CURRENT identifiers are processed by Batch CD-Search only if the option to include retired sequences (originally called "search ID1 for unknown identifers") is activated. If it is deactivated, any non-current identifiers in your query list will be ignored by Batch CD-Search and results will be returned only for the current identifiers. The Batch CD-Search output will flag each non-current identifier with the message, "Warning: this sequence record may be obsolete or preliminary."
If an identifier is not found in either the live Entrez Protein database or in the archival database
, it will be considered INVALID and will be ignored by the Batch CD-Search program. Invalid identifiers will not appear in the sample data table of the preliminary job summary page, or in dowloaded output data files. They will, however, appear in the "navigate results" menu of the graphical display of search results, listed as " Query #N - XXXXXXXX(invalid)" in greyed out font. - Protein sequences




- FASTA format or bare sequence data -- Query protein sequences can also be entered into Batch CD-Search in FASTA format or simply as bare sequence data (single letter code). In both cases, the protein data must end with an empty line (i.e., the sequence data must be followed by two consecutive newline characters (\n\n) to indicate the end of data). For FASTA format, the ">" character must appear as the first character of the definition line (defline) of FASTA formatted sequence, otherwise the defline may be parsed as an identifier and will therefore be interpreted as a different query from the sequence data.
- Maximum input




- Up to 4,000 protein sequences and/or identifiers can be input into Batch CD-Search, either through the web interface or through scripting. Requests containing more than 4000 queries will be rejected as peak usage of this shared resource has increased significantly and has impinged service availability.
- Adjust search options




- Search mode




- automatic -- The Batch CD-Search program chooses to retrieve either precalculated or live search results for each separate item in the query list, depending on the nature of the item. For example, if the query item is a valid UID, the program will always try to retrieve the precalculated search result. If that fails, the program will then try a live RPS-BLAST search. However, if sequence data are submitted explicitly (as FASTA or base sequence), the program goes directly to live search. (Note that the "automatic" search mode uses the default search parameters; if you would like to change the parameters from their default settings, use the "live search only" mode.)
- precalculated only -- The Batch CD-Search program will retrieve only precalculated data for sequence identifiers in your query list. If precalculated data are not available for a given sequence identifier, no search result will be returned for that item. Note that this search mode works only for input that was entered as sequence identifiers; if your input was sequence data, no results will be returned. (Also note that the "precalculated only" mode uses the default search parameters; if you would like to change the parameters from their default settings, use the "live search only" mode.)
- live search only -- The Batch CD-Search program will do a live PRS-BLAST search for every item in the input list, whether the item is a sequence identifier or sequence data. This mode also allows you to change search parameters from their default settings.
- database selection -- currently, Batch CD-Search is offered with the search databases listed below. As mentioned in the introduction to adjusting search parameters, the Batch CD-Search system uses the "automatic" search mode by default, which searches against the "CDD" database. If you change the database against which you want to search, the search mode will automatically change to "live search only." (If you change the search mode back to "automatic," the search parameters will be reset to their default values.)
- CDD - this is a superset including NCBI-curated domains and data imported from Pfam, SMART, COG, PRK, and TIGRFAMs. It is the default database for searches.
- NCBI_Curated - NCBI-curated domains, which use 3D-structure information to explicitly to define domain boundaries, aligned blocks, and amend alignment details, and which aim to provide insights into how patterns of residue conservation and divergence in a family relate to functional properties.
- Pfam - a mirror of a recent Pfam-A database of curated seed alignments. Pfam version numbers do change with incremental updates. As with SMART, families describing very short motifs or peptides may be missing from the mirror. An HMM-based search engine is offered on the Pfam site.
- SMART - a mirror of a recent SMART set of domain alignments. Note that some SMART families may be missing from the mirror due to update delays or because they describe very short conserved peptides and/or motifs, which would be difficult to detect using the CD-Search service. You may want to try the HMM-based search service offered on the SMART site. Note also that some SMART domains are not mirrored in CD because they represent "superfamilies" encompassing several individual, but related, domains; the corresponding seed alignments may not be available from the source database in these cases. Note also that SMART version numbers do not change with incremental updates of the source database (and the mirrored CD-Search database).
- PRK - "PRK," short for Protein Clusters, is an NCBI collection of related protein sequences (clusters) consisting of Reference Sequence proteins encoded by complete prokaryotic and chloroplast plasmids and genomes. It includes both curated and non-curated (automatically generated) clusters.
- TIGRFAMs - a mirror of a recent TIGRFAMs set of domain alignments.
- COG - a mirror of the current COG database of orthologous protein families focusing on prokaryotes. Seed alignments have been generated by an automated process. An alternative search engine, "Cognitor", which runs protein-BLAST against a database of COG-assigned sequences, is offered on the COG site.
- KOG - a eukaryotic counterpart to the COG database. KOGs are not included in the CDD superset, but are searchable as a separate data set.
More information about each database is provided in the section on "Where does CDD content come from?" and the version number (as available) of each source database is provided in the CDD News page.
- expect value (E-value) threshold -- is a parameter that describes the number of hits one can "expect" to see by chance when searching a database of a particular size . The E-value setting can be modified to adjust the statistical significance threshold used for reporting matches against PSSMs in the database. False positive results should be very rare with the default setting of 0.01 (use a more conservative, i.e. lower, setting for more reliable results). Results with E-values in the range of 1 and above should be considered putative false positives. Additional information about E-value is available in the Glossary of the NCBI Handbook and in the BLAST help document .
- composition-corrected scoring, which is employed by RPS-BLAST version 2.2.28 (March 19, 2013) and up, abolishes the need to mask out compositionally biased regions in query sequences. This option is on by default .
Note: In general, when composition-corrected scoring is on, the low complexity filter should be turned off. However, it is possible to have both options on at the same time (to filter false-positives that slip through the cracks of the composition-correction), or off at the same time (to find more distant relatives for compositionally biased queries), if desired. - low complexity filter -- filters query sequences for compositionally biased regions. These regions are flagged as such and largely ignored during the search phase if filtering is turned ON (the default setting is OFF).
Note: In general, when the low complexity filter is turned on, the composition-corrected scoring should be turned off. However, it is possible to have both options on at the same time (to filter false-positives that slip through the cracks of the composition-correction), or off at the same time (to find more distant relatives for compositionally biased queries), if desired.If the low-complexity filter is turned on and compositially biased regions are detected, they are shown in the Batch CD-Search graphical display as cyan regions in the bar graphic that represents the query sequence, as illustrated below. More information about the low complexity filter is also available in the BLAST help document.
- If the low-complexity filter was ON for the search, the compositionally biased regions were NOT USED in the search against the domain database and are shown as SOLID cyan blocks. (As an example, open the default CD-Search results for P14780, GI 269849668, with filtering turned ON.) However, those regions may still overlap with or be included in a domain footprint and the pair-wise alignment generated by RPS-BLAST.

- If the low-complexity filter was turned OFF for the search, the compositionally biased regions were USED in the search and are shown as blocks OUTLINED in cyan. (As an example, open the CD-Search results for P14780, GI 269849668, with filtering turned OFF.) Please keep in mind, however, that compositionally biased regions can cause inaccurate annotation of the query sequence.

- If the low complexity filter DID NOT DETECT any compositionally biased regions in the query sequence, then it is displayed as a plain grey bar (with no cyan regions), as shown in the illustrations of the sample concise display and full display of CD-Search results.
- maximum number of hits -- limits the size of the hit list produced by CD-Search. Typically, for average sized proteins, the number of expected domain-hits is small and the default setting of 500 should be more than sufficient.
- Include retired sequences (originally called "Search ID1 for unknown IDs")




- This option searches the backend archival database for any protein sequence identifiers in your input list that are not recognized as being current in the live Entrez Protein database. If these sequence identifiers are found in the archival database, the Batch CD-Search program will retrieve results for them, even if they have been deprecated. This option is activated by default. If it is deactivated, any non-current identifiers in your query list will be ignored by Batch CD-Search and results will be returned only for the current identifiers. (See the section on UID validation for additional details.)
- Optional job title




- An arbitrary string can be specified as a title for a particular search job, with a maximum of 256 characters. (If a longer strong is provided, it will be truncated.) The job title is not used in any way by the search engine. It is therefore totally optional but recommended for easy identification of search results, especially when multiple jobs are submitted simultaneously.
- E-mail address(es) to receive notification when job is done




- You can provide one or more email addresses here in order to receive notification when the search job is done. Multiple email addresses must be separated by commas. The title of the job, if assigned, will appear in the subject line.
- Retrieve a previous search result by entering its search identifier




- When a Batch CD-Search is successfully submitted, a unique, randomly generated identifier, or "Search ID," is assigned to identify the search (for example, QM2-qcdsearch-xxxxxxxxxxx) . The Search ID can be used to retrieve the search status/results for up to 2 days after the search was first run. To do this, enter the Search ID in the "Retrieve a previous search" text box on the Batch CD-Search page and click the "Retrieve" button. More details about Search IDs are provided in a separate part of this document.
By default, the "search mode" is set to "automatic." This mode automatically applies the search parameters that were used to generate precalculated results for all sequences in the NCBI Protein database, and provides the fastest way of obtaining Batch CD-Search results.The search mode will automatically change to "live search only" if you change the database against which you want to search, use a less stringent expect value, apply composition-corrected scoring, or apply the low-complexity filter.
If you change the search mode back to "automatic," the search parameters will be reset to their default values.
Note: Modifying the "maximum number of hits" or activating/deactivating the option to "include retired sequences" does not change the search mode. Instead, it just filters your search results as you have specified.
More details about search mode and each of the other parameters are provided below:
![]()
![]()
![]()
![]()
| job summary, statistics, & sample data table | download options: data type (target data) , data mode | graphical display |
- Job summary




- Search completed successfully -- After a Batch CD-Search has been successfully run (see job status codes), a preliminary web page is displayed with the message, "search completed successfully." This indicates the complete search results have been compiled into a temporary database, which serves as a master data structure from which you can choose to download data (domain hits, alignment details, or features) or view the results graphically. Once you select the desired download or graphical viewing option, the program extracts the specified output from the master data structure and presents it as a text file or web display. The master data structure remains available to you for up to 2 days after the search was first run. Be sure to save the search ID in order to retrieve the results over that time period, either through the "retrieve a previous search" text box on the Batch CD-Search home page or through scripted data downloads using a Web API.
- Statistics




- Sample data table -- The job summary page that appears after a Batch CD-Search has been successfully completed shows the concise list of hits on the first ten protein sequences from your query list. To see the results for all the proteins in your query list, use the dowload options or the graphical display.
Explanations of the column headers that appear in the sample data table are provided in the domain hits section of this help document and are also accessible by clicking on the column headers in the sample output file.
Note: If there are any old/deprecated sequence identifiers in the query list, the "Sample data" table will display a warning beside those identifiers that indicates "this sequence record may be obsolete or preliminary." Nevertheless, conserved domain search rsults will be returned for those identifiers as long as the option to include retired sequences (originally called "search ID1 for unknown identifers") was activated for the search. If any of the identifiers in the query list were invalid, they will not appear in the sample data table or in the output data files generated by the dowload options. (Invalid identifiers will, however, appear in the "navigate results" menu of the graphical display of search results, listed as " Query #N - XXXXXXXX(invalid)" in greyed out font.)
- Download options




- Data type (target data)




- Domain hits -- A list of conserved domain models, from the database you selected to search, that have statistically significant hits to the protein sequences in your query list. (An example is provided in a separate file.)




- Alignment details -- Data files that list the pairwise alignment details between each query protein sequence from the input file and the consensus sequence from each domain model or superfamily that had a hit to the sequence. You can choose to download the alignments for the concise, standard, or full list of hits.




- Features -- Data files that list the conserved features/sites, such as catalytic residues, binding sites, or motifs, found on the protein query sequences.




- Data mode




- Concise -- The Concise result is the default output for Batch CD-Search and lists only the best scoring domain model, as available for each region on the query sequence, in each of three hit types: specific hits, the superfamily to which the highest-ranking hit belongs, and multi-domain models.
If CD-Search finds both specific and non-specific hits for a region of a protein query sequence, only the highest ranking specific hit and its superfamily will be shown. If CD-Search finds only non-specific hits for a region of a protein query sequence, only the superfamily to which the hits belong will be shown, but not the non-specific hits themselves (illustrated example). The latter are provided only in the full display.
- Standard -- The Standard result lists the best scoring domain model from each source database, as available for each region on the query sequence In some cases, two NCBI-curated models might be shown for a given region of a protein, if the immediate parent of the highest ranking NCBI-curated conserved domain model is also in the search results. (A separate section of this document provides more information about domain family hierarchies.)
- Full -- The Full result lists all domain models, as available for each region on the query sequence, that meet or exceed the RPS-BLAST threshold for statistical significance (i.e., the E-value cutoff). The hit types can include specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models (illustrated example). Hits are ranked by E-value, although NCBI-curated models are ranked ahead of other hits to the region if their E-value exceeds a threshold of 1e-05.
- Additional options -- For downloads of domain hits or alignment details, you can also choose to (a) retrieve superfamily hits only; (b) include the query defline (query protein sequence definition line), and/or (c) include the domain defline (short name of the domain). For downloads of features, only option (b) is available.
- Browse Results/Graphical display




- Left Panel offers controls that allow you to select any individual protein(s) from your query list for which you want to graphically view domain annotations, or to download the complete search results.
- Navigate results -- The left side of the browser window contains a "Navigate results" box that lists each query sequence from your original input list. The sequences are shown in the format Q#N - XXXXXXXX, where Q#N is the query number and XXXXXXXX is either the sequence identifier, the first 15 characters of the FASTA definition line, or the first 15 amino acids of bare sequence data. Click on any query sequence to view a graphical display of its domain hits and features. If you would like to select multiple query sequences from the list, use the CTRL or SHIFT keys while clicking on the desired sequences.
- Compact Mode -- The "Compact mode" option in the "Navigate results" box displays the domain architecture of each query sequence on a single line. This display type is particularly useful if you select two or more query proteins from the list and want to compare their domain architectures. (As noted above, you can use the CTRL or SHIFT keys while clicking on the query proteins that are listed in the "Navigate results" box, if you would like to select multiple sequences from that list.)
- Note: If your input listed sequence identifiers and some of those identifiers were invalid, their query numbers and identifiers appear as greyed out text in the format " Query #N - XXXXXXXX(invalid)" in the "navigate results" menu. (Invalid identifiers and their query numbers will not, however, appear in downloaded data files.)
- Download data -- The "Download data options" beneath the "navigate results" box are the same as those displayed on the job summary page.
- Statistics -- presents the same statistics as those displayed on the job summary page.
- Right Panel presents a graphical display of the conserved domain domain footprints on any individual protein sequence from your query list.
- The initial display shows the domain footprints on the first query sequence. Use the "Navigate results" box to select any other protein query sequence from your input list. A concise display (illustrated example) of domains is shown by default. If conserved features/sites have been found as well, they will appear as small triangles (illustrated example).
- The "Show functional sites" and "View: Concise/Standard/Full display" controls at the right hand edge of the graphic display allow you to turn the feature annotations on or off, and to select the desired level of detail in the display.
- Mouseover any domain footprint to view a pairwise alignment of the query sequence to the consensus sequence of the domain model.
- If the footprint represents a superfamily, click on the footprint to open the corresponding superfamily record, which in turn lists the various domain models encompassed within it.
- If the footprint is a specific hit (visible in both the concise and full display of search results) or a non-specific hit (visible in only the full display of search results), you can click on the footprint to view the query sequence embedded in the multiple sequence alignment for the domain model.
- Mouseover any small triangle to view information about the conserved feature/site that has been mapped to the query sequence.
- Click on the triangle to view additional details about the feature, including a multiple sequence alignment of your query sequence and the protein sequences used to curate the domain model, where hash marks (#) above the aligned sequences show the location of the conserved feature residues.
- If a 3D structure is included among the evidence used to annotate the feature, the details page will show a thumbnail image, which provides an approximate view of the feature's location in 3 dimensions and allows you to open an interactive 3D structure view in the free Cn3D program.
- What is the significance of the domain colors/shapes?
- Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other. (Because the number of superfamilies exceeds the number of different color/shape combinations, some of the superfamilies share the same display style. It should be rare, though, to encounter such pairs in the same domain architecture.)
- The color/shape combination remains stable for a given superfamily cluster ID, and is consistent across all NCBI tools that show conserved domain footprints on protein sequences (i.e., CD-Search, Batch CD-Search, and CDART).
- If the cluster ID for a superfamily changes (due to significant changes in the superfamily composition), the color/shape combination may also change.
- "Specific hits" are shown as bright colors, with full saturation. The "superfamily" to which a "specific hit" belongs is shown as a pastel color, with lower saturation.
- If there are no specific hits to a region of a protein query sequence, then the "Concise display" will show only the superfamily. Regardless whether there are specific hits or not, "Standard display" will show the superfamily plus highest ranking hit from each source database, and the "Full display" will show the all the domain models in each hit type (specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models).

Search ID When a search is started, a unique, randomly generated search ID is returned to identify the query and the master data structure that holds the complete set of results retrieved by the search. (The program later uses that master data structure to extract whatever subset of information the user has requested and constructs the final output, depending on the parameters you specify.) The Search ID starts with "QM2-qcdsearch-" as a signature and is followed by a randomly generated hex number (xxxxxxxxxxx), for example:
QM2-qcdsearch-xxxxxxxxxxxWhen specific subset of output data (for example, domain hits, alignments, or features) are requested, a new session is launched to construct the desired output and a second hex number (yyyyyyyyyyy) is returned to identify it. This number can be attached to the end of a search ID to form a complete data request ID, for example:QM2-qcdsearch-xxxxxxxxxxx-yyyyyyyyyyyBoth forms of search ID are valid as input for the "retrieve a previous search" text box on the Batch CD-Search home page and for scripted data downloading/handling (using the cdsid parameter). The Search ID can be used to retrieve the search status/results for up to 2 days after the search was first run.Data source The database of conserved domains (or more technically, of PSSMs) against which your list of protein query sequences was searched. If your search mode was "automatic" or "precalculated only," then your queries were searched against the default CDD database. If your search mode was "live search only," this line reports the database you selected.
E-value cutoff The statistical significance threshold used for reporting matches against PSSMs in the database. If your search mode was "automatic" or "precalculated only," then the default E-value of 0.01 was applied. If your search mode was "live search only," this line reports the database you selected.
Composition-corrected scoring This line indicates whether composition-corrected scoring was used. The values reported in this line can be either "0" (off) or "1" (on, which is the default setting). The filter is on when the search mode is "automatic" or "precalculated only," but can be turned off if desired when the mode is "live search only." Important note: Composition-corrected scoring should not be used when the low-complexity filter is on.
Low-complexity regions This line indicates whether query sequences were filtered for low complexity regions. The values reported in this line can be either "Filtered" or "Not Filtered (default)." The filter is off when the search mode is "automatic" or "precalculated only," but can be turned on if desired when the mode is "live search only." Important note: The low-complexity filter should not be used when composition-corrected scoring is on.
Maximum aligns The maximum number of domain hits that will be reported for any individual protein from your query list. If your search mode was "automatic" or "precalculated only," then the default setting of 500 was applied. If your search mode was "live search only," this line reports the setting you selected.
Run time The amount of time the it took to execute the search, starting from the time it was pulled from the job queue ending at the time at which the master data structure (raw search results from which various outputs can be generated) has been constructed. The run time does not include the time during which the query is in the queue, or the time that is required by the program to save/retrieve from cache, construct/format output data, etc.
Total queries parsed The total number of protein sequence identifiers and/or sequence data strings detected by the program in your input list.
Total valid queries The total number of valid protein sequence identifiers and sequence data strings found in your query list.
Queries with no domain hits The total number of valid queries for which no domain hits were found.
Total domains found The total number of conserved domains found in the full results (i.e., the complete set of domain models from CDD that were hits to any query sequence, with each database record counted only once regardless of how many query sequences that model hit).
Total clusters found The total number of CDD superfamily clusters found in the full results (i.e., the complete set of CDD superfamilies to which the conserved domain models (referenced in "total domains found") belong, with each superfamily record counted only once).
Total specific features found The total number of conserved features/sites that were mapped onto the set of query sequences from specific hits. Each conserved feature/site is counted only once, regardless of how many query sequences on which it was found.
Total generic features found The total number of conserved features/sites that were mapped onto the set of query sequences from non-specific hits, because those non-specific hits belong to a superfamily whose representative is an NCBI-curated domain that has such annotations.
| data type: domain hits, alignment details , features |
| data mode: concise, standard, full |
The download options accessible on the web interface allow you to specify which data elements (e.g., domain hits, alignment details, or features) you would like to extract from the master data structure into the output files, and at what level of comprehensiveness (concise, standard, or full results). These options are also available for scripted data downloads and can be specified with the valid parameters.
Format: tab delimited table that lists the following information for each protein sequence in your query list:
Q#N - XXXXXXXX Query number: The ordinal number (N) of the query sequence from your original input list. The query number is recorded as Q#N - XXXXXXXX, where XXXXXXXX is either the sequence identifier, the first 15 characters of the FASTA definition line, or the first 15 amino acids of bare sequence data . Note: If your input listed sequence identifiers and some of those identifiers were invalid, their query numbers and identifiers will be missing from the output file, but can be seen in the "navigate results" menu of the graphical display of search results, if desired. For example, if your input file contained four sequence identifiers and the third one was invalid, the output file will show the results for Q#1, Q#2, and Q#4. The invalid query (Q#3) can be seen, however, as greyed out text in the format " Query #3 - XXXXXXXX(invalid)" in the graphical display.
Hit type CD-Search results can include hit types that represent various confidence levels (specific hits, non-specific hits) and domain model scope (superfamilies, multi-domains). They can be seen in both the Concise display and Full display, except for non-specific hits, which are shown only in the Full Display. PSSM-ID A PSSM ID is the unique identifier for a domain model's position-specific scoring matrix (PSSM). If a domain model's PSSM changes in any way as a result of updates to its multiple sequence alignment, it receives a new PSSM ID. Each superfamily record in the Conserved Domain Database also has a PSSM ID, which refers to the specific set of conserved domain PSSM IDs that comprise the superfamily, rather than to an actual position-specific scoring matrix for the overall superfamily. more... From..To The range of amino acids in the query protein sequence to which the domain model aligns. (Note: If the alignment found by RPS-BLAST omitted more than 20% of the CD's extent at either the n- or c-terminus or both, the partial nature of the hit is indicated in the "Incomplete" column of the hit table. Partial hits can also be spotted in the graphical display as domain model cartoons with jagged edges (illustrated example).) E-value The expect value, or E-value, indicates the statistical significance of the hit as the likelihood the hit was found by chance. more... Bit Score The value S' is derived from the raw alignment score S in which the statistical properties of the scoring system used have been taken into account. Because bit scores have been normalized with respect to the scoring system, they can be used to compare alignment scores from different searches. (A bit score is defined in the NCBI Handbook glossary and Field Guide glossary.) Accession The accession number of the hit, which can either be a domain model or a superfamily cluster. (If the hit is a domain model, then the accession number (cl*) of the superfamily cluster to which it belongs is listed in the "Superfamily" column of the output file.) Short name The short name of a conserved domain, which concisely defines the domain. For example, "Voltage gated ClC" is the short title of the NCBI-curated conserved domain model for the voltage gated chloride channel (cd00400). Incomplete If the hit to a conserved domain is partial (i.e., if the alignment found by RPS-BLAST omitted more than 20% of the CD's extent at either the n- or c-terminus or both), this column will be populated with one of the following values:
N: incomplete at the N-terminus
C: incomplete at the C-terminus
NC: incomplete at both the N-terminus and C-terminus
(the aligmnent found by RPS-BLAST omitted
more than 40% of the CD's total extent)
If the hit to a conserved domain is complete, then this column will be populated with a dash (-).
(Note: Partial hits can also be spotted in the graphical display as domain model cartoons with jagged edges (illustrated example).)Superfamily This column is populated only for domain models that are specific or non-specific hits, and it lists the accession number of the superfamily to which the domain model belongs. (If the hit is to a superfamily itself, then this column is simply populated with a dash because the superfamily accession is already listed in the preceding "Accession" column.)
Format options:
Click on any one of the first three formats above to read more about it. The BLAST text format is described below.The BLAST text format for downloading alignment details from Batch CD-Search results displays a pairwise alignment between the protein query sequence and the consensus sequence from each domain model and/or superfamily that had a hit to the sequence. Exact matches are marked by a pipe symbol ("|") between query and database sequence. As an example, below is an excerpt of the BLAST text formatted alignment details for domain hits on NP_000240 (GI:4557757). Click on the example below to open the complete sample file, representing the concise results for that query sequence as of 01 November 2010:
Format: tab delimited table that lists the following information for each protein sequence in your query list:
Q#N - XXXXXXXX Query number: The ordinal number (N) of the query sequence from your original input list. The query number is recorded as Q#N - XXXXXXXX, where XXXXXXXX is either the sequence identifier, the first 15 characters of the FASTA definition line, or the first 15 amino acids of bare sequence data . Note: If any query numbers are missing from the output file, that indicates that either: (a) no features were found on those protein sequences, or (b) the sequence identifiers were invalid. (Invalid sequence identifiers can be seen in the "navigate results" menu of the graphical display of search results, if desired. For example, if your input file contained four sequence identifiers and the third one was invalid, the output file will show the results for Q#1, Q#2, and Q#4. The invalid query (Q#3) can be seen, however, as greyed out text in the format " Query #3 - XXXXXXXX(invalid)" in the graphical display.)
Type The feature type can be either: specific: conserved features/sites that were mapped onto the set of query sequences from specific hits.
generic: conserved features/sites that were mapped onto the set of query sequences from non-specific hits, because those non-specific hits belong to a superfamily whose representative is an NCBI-curated domain that has such annotations.
Title The brief name of the conserved feature/site, for example, "active site," "catalytic tetrad," "Ca2+ binding site," etc. Coordinates A comma delimited list of the single letter amino acid codes and their positions on the query sequence, indicating which residues in the query protein align to the conserved feature/site annotated on the domain model. For example: D50,Y55,K84,H117 Complete size The total number of residues in the conserved feature/site that has been annotated on the domain model. Mapped size The number of residues in the query protein sequence that match residues in the conserved feature/site that was annotated on the domain model. Source domain The PSSM ID of the domain model on which the conserved feature/site was annotated.
On the Batch CD-Search job summary page, a "Browse Results" button above the sample data table allows you to view the results graphically. The button opens a separate browser window that shows the domain footprints, alignment details, and conserved features on any individual query sequence. The browser window is divided into two parts:
![]()
![]()
![]()
![]()
| HTTP GET or POST requests | maximum input |search ID |
| submit: base URL, parameters, examples | check status | retrieve |
| sample PERL script for HTTP POST operations: input, PERL script, sample run, output |
If invoked with parameters, Batch CD-Search can be used as an interface for scripted data downloading/handling. A query can be submitted as an HTTP GET or an HTTP POST request.An HTTP GET request is submitted as a URL and can contain a maximum of approximately 1000 characters. There is no character limit on the length of an HTTP POST request, but there is a limit of 4,000 protein sequences and/or identifiers in a single Batch CD-Search request.
The program retrieves output data in two steps: First it does the search and collects all available information from the search results into a master data structure, then it extracts the subset of information the user has requested and constructs the final output.
When a search is started, a unique, randomly generated search ID is returned to identify the query and the master data structure that holds the complete set of results retrieved by the search. (The program later uses that master data structure to extract whatever subset of information the user has requested and constructs the final output, depending on the parameters you specify.) The Search ID starts with "QM2-qcdsearch-" as a signature and is followed by a randomly generated hex number (xxxxxxxxxxx), for example:QM2-qcdsearch-xxxxxxxxxxxWhen specific subset of output data (for example, domain hits, alignments, or features) are requested, a new session is launched to construct the desired output and a second hex number (yyyyyyyyyyy) is returned to identify it. This number can be attached to the end of a search ID to form a complete data request ID, for example:QM2-qcdsearch-xxxxxxxxxxx-yyyyyyyyyyyBoth forms of search ID are valid as input (using the cdsid parameter) for scripted data downloading/handling. The Search ID can be used to retrieve the search status/results for up to 2 days after the search was first run.
- Submit




- base URL:




- valid parameters:




- examples of URLs for HTTP GET requests:




- Check status




- enter the Search ID (QM2-qcdsearch-xxxxxxxxxxx or QM2-qcdsearch-xxxxxxxxxxx-yyyyyyyyyyy), which is valid for up to 2 days after the search was first run. For example:
- Retrieve




- search summary page -- If you entered a search ID in the format QM2-qcdsearch-xxxxxxxxxxx (as in the check status sample URL) and the search you are checking on was already completed successfully, the program will display the default search system output on a web page.
- specific target data -- If you entered a search ID in the format QM2-qcdsearch-xxxxxxxxxxx-yyyyyyyyyyy and the search you are checking on was already completed successfully, the program will return the specific target data you requested.
- Sample PERL script for HTTP POST operations:




- sample input file -- An input file can contain protein sequence identifiers and/or sequence data. The following example (with filename "samplefile.in") contains a mixture of GIs, accessions, and raw sequence data:
- sample PERL script -- the sample script below (with filename "bwrpsb.pl") includes statements to validate input, set default values and parameters, submit the search, check status of the job, and retrieve the results:
- sample run of PERL script -- At your computer's prompt ($), run the PERL script with a command such as the following:




- output -- The Batch CD-Search output file that is returned for an HTTP POST request will contain conserved domain hits, alignment details, or features, as you specified. An example output file, from the sample PERL script above, opens in a separate window and contains a tab-delimited table of conserved domain hits on the protein query sequences.




https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi?
to check the status of, or retrieve results for, a previous search: cdsid Specify the search ID string, for example, "cdsid=QM2-qcdsearch-xxxxxxxxxxx" if you want to check the status of a previous search. Include a second hex number, for example, "cdsid=QM2-qcdsearch-xxxxxxxxxxx-yyyyyyyyyyy" if you want to retrieve specific output data that were previously requested for the search. (Both forms of search ID are valid for up to 2 days after the search was first run.) If you want to retrieve a different subset of output data for a previously run search, specify the search ID string, for example, "cdsid=QM2-qcdsearch-xxxxxxxxxxx," plus any of the output parameters described below. to run a new search: db Specify the name of the database. Allowable values include: "cdd," "pfam," "smart," "tigrfam," "cog," "kog". This parameter only applies if the search mode (smode) is live. If the search mode is set to precalculated or automatic, then the default CDD database is searched. smode Specify the desired search mode: "auto" (automatic), "prec" (precalculated only), or "live" (live) useid1 "true"/"false", this parameter specifies whether the program should search the backend archival database for any protein sequence identifiers in your input list that are not recognized as being current in the live Entrez Protein database. If these sequence identifiers are found in the archival database, the Batch CD-Search program will retrieve results for them, even if they have been deprecated. This option is activated by default. If it is deactivated, any non-current identifiers in your query list will be ignored by Batch CD-Search and results will be returned only for the current identifiers. (See the section on UID validation for additional details.) compbasedadj "0"/"1" , this parameter specifies whether the program will employ composition-corrected scoring. The possible values are: 0: NoCompositionBasedStats (composition-based statistics turned off)
1: CompositionBasedStats (composition-based statistics turned on (default))filter "true"/"false", this parameter specifies whether the program will filter out compositionally biased regions from the query sequences. The default setting is "false".
Note: In general, if the low complexity filter is turned on, the composition-corrected scoring should be turned off. However, it is possible to have both options on at the same time (to filter false-positives that slip through the cracks of the composition-correction), or off at the same time (to find more distant relatives for compositionally biased queries), if desired.queries Specify the query proteins, either as unique identifiers or as sequence data. Refer to the "input" section of this document for rules. evalue Floating point number, specifies the expect value (E-value) cut-off, which adjusts the statistical significance threshold used for reporting matches against PSSMs in the database. This parameter only applies if the search mode (smode) is live. If the search mode is set to precalculated or automatic, then the default E-value of 0.01 is applied. maxhit Integer, specifies the maximum number of hits to return for each protein in the query list. This parameter only applies if the search mode (smode) is live. If the search mode is set to precalculated or automatic, then the default maxhit number of 500 is applied. to specify the desired output for a new or previous search: tdata Specify the data type (target data) desired in the output. Allowable values are: "hits" (domain hits), "aligns" (alignment details), or "feats" (features). alnfmt If you have specified tdata=aligns (alignment details), then you can use the "alnfmt" parameter to specify the desired download format. Allowable values are: "asn", "xml", or "json". dmode Specify the data mode desired in the output. Allowable values are:
"rep" (highest scoring hit, for each region of the query sequence, as shown in the concise results), or
"std" (best-scoring hit from each source database, for each region of the query sequence, as shown in the standard results), or
"full" (the complete set of hits in the full results).
(The value "all" is still allowed and previously was used to display the full result, but now displays the standard result, since Feb. 12, 2014, when the standard mode became available as a new display option.)qdefl "true"/"false", this parameter specifies whether to include definition lines for the query proteins in the output. cddefl "true"/"false", this parameter specifies whether to include the titles of conserved domains in the output.
Sample#1: Submit query proteins with sequence identifiers 116863, 122, 1065303, and 109389365; check the backend archival database for invalid sequence identifiers; and return a concise list (which is the default setting, as no dmode parameter is specified) of domain hits in the output:https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi?queries=116863%0A122%0A1065303%0A109389365&useid1=true&tdata=hits
Sample #2: For a previously run search with cdsid=QM2-qcdsearch-xxxxxxxxxxx, retrieve the alignment details (tdata=aligns) in XML format (alnfmt=xml) for the full results (dmode=all):https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi?cdsid=QM2-qcdsearch-xxxxxxxxxxx&tdata=aligns&alnfmt=xml&dmode=all
Note: In addition to the sample HTTP GET requests above, some sample PERL scripts for HTTP POST requests are provided in a subsequent section of this document.
Note that the sample search ID above will not work at this time because a search ID is valid for only 2 days after the search was first run. It is provided here only as an example.https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi?cdsid=QM2-qcdsearch-604A3735FD77DDEF
If a search is still in progress, the program will return one of the status codes below. If a search has been successfully completed, the program will retrieve a search summary page or specific target data, depending on the search ID format you provide in the URL, as noted in the next section.
| input | script | sample run | output |
samplefile.in122 116863 8CAT_A P90895 MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQ YMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDLAARTVESRQAQDLARSYGIP YIETSAKTRQGVEDAFYTLVREIRQHKLRKL
bwrpsb.pl#!/usr/bin/perl -w use strict; use LWP::UserAgent; use Getopt::Std; ############################################################################### # URL to the Batch CD-Search server ############################################################################### my $bwrpsb = "https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi"; ############################################################################### # read list of queries and parameters supplied; queries specified in list piped # from stdin ############################################################################### my @queries = <STDIN>; my $havequery = 0; ############################################################################### # do some sort of validation and exit if only invalid lines found ############################################################################### foreach my $line (@queries) { if ($line =~ /[a-zA-Z0-9_]+/) { $havequery = 1; } } if ($havequery == 0) { die "No valid queries!\n"; } ############################################################################### # set default values ############################################################################### my $cdsid = ""; my $cddefl = "false"; my $qdefl = "false"; my $smode = "auto"; my $useid1 = "true"; my $maxhit = 250; my $filter = "true"; my $db = "cdd"; my $evalue = 0.01; my $dmode = "rep"; my $clonly = "false"; my $tdata = "hits"; ############################################################################### # deal with command line parameters, change default settings if necessary ############################################################################### our($opt_d, $opt_e, $opt_F, $opt_b, $opt_t, $opt_s, $opt_a, $opt_q); getopts('d:e:F:b:t:s:a:q'); if ($opt_d) { $db = $opt_d; print "Databast option set to: $db\n"; } if ($opt_e) { $evalue = $opt_e; print "Evalue option set to: $evalue\n"; } if ($opt_F) { if ($opt_F eq "F") { $filter = "false" } else { $filter = "true"; } print "Filter option set to: $filter\n"; } if ($opt_b) { $maxhit = $opt_b; print "Maxhit option set to: $maxhit\n"; } if ($opt_t) { $tdata = $opt_t; print "Target data option set to: $tdata\n"; } if ($opt_s) { $clonly = "true"; print "Superfamilies only will be reported\n"; } if ($opt_a) { $dmode = "all"; print "All hits will be reported\n"; } if ($opt_q) { $qdefl = "true"; print "Query deflines will be reported\n"; } ############################################################################### # submitting the search ############################################################################### my $rid; { my $browser = LWP::UserAgent->new; my $response = $browser->post( $bwrpsb, [ 'useid1' => $useid1, 'maxhit' => $maxhit, 'filter' => $filter, 'db' => $db, 'evalue' => $evalue, 'cddefl' => $cddefl, 'qdefl' => $qdefl, 'dmode' => $dmode, 'clonly' => $clonly, 'tdata' => "hits", ( map {; queries => $_ } @queries ) ], ); die "Error: ", $response->status_line unless $response->is_success; if($response->content =~ /^#cdsid\s+([a-zA-Z0-9-]+)/m) { $rid =$1; print "Search with Request-ID $rid started.\n"; } else { die "Submitting the search failed,\n can't make sense of response: $response->content\n"; } } ############################################################################### # checking for completion, wait 5 seconds between checks ############################################################################### $|++; my $done = 0; my $status = -1; while ($done == 0) { sleep(5); my $browser = LWP::UserAgent->new; my $response = $browser->post( $bwrpsb, [ 'tdata' => "hits", 'cdsid' => $rid ], ); die "Error: ", $response->status_line unless $response->is_success; if ($response->content =~ /^#status\s+([\d])/m) { $status = $1; if ($status == 0) { $done = 1; print "Search has been completed, retrieving results ..\n"; } elsif ($status == 3) { print "."; } elsif ($status == 1) { die "Invalid request ID\n"; } elsif ($status == 2) { die "Invalid input - missing query information or search ID\n"; } elsif ($status == 4) { die "Queue Manager Service error\n"; } elsif ($status == 5) { die "Data corrupted or no longer available\n"; } } else { die "Checking search status failed,\ncan't make sense of response: $response->content\n"; } } print "===============================================================================\n\n"; ############################################################################### # retrieve and display results ############################################################################### { my $browser = LWP::UserAgent->new; my $response = $browser->post( $bwrpsb, [ 'tdata' => $tdata, 'cddefl' => $cddefl, 'qdefl' => $qdefl, 'dmode' => $dmode, 'clonly' => $clonly, 'cdsid' => $rid ], ); die "Error: ", $response->status_line unless $response->is_success; print $response->content,"\n"; }
$ ./bwrpsb.pl < samplefile.in Search with Request-ID QM2-qcdsearch-5F8CF46DDF26149C-5F8CF46DDF26149C started. ......Search has been completed, retrieving results ..
![]()
![]()

Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020 Jan 8;48(D1):D265-D268. doi: 10.1093/nar/gkz991. (Epub 2019 Nov 28.) [PubMed PMID: 31777944] [Full Text at Oxford Academic] 
![]()
![]()

Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W327-31. [Full Text at Oxford Academic] Note: If using Batch CD-Search, please also cite a second article, which discussed the launch of that resource:
[Full Text at Oxford Academic] [Full text in PubMed Central]
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Deweese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011 Jan;39(Database issue):D225-9. doi: 10.1093/nar/gkq1189. Epub 2010 Nov 24. [PubMed PMID: 21109532]
![]()
![]()
How to Add New Line in Domain_9 Summary
Source: https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml
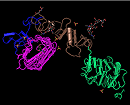
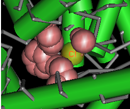



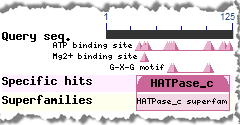



0 Response to "How to Add New Line in Domain_9 Summary"
Post a Comment